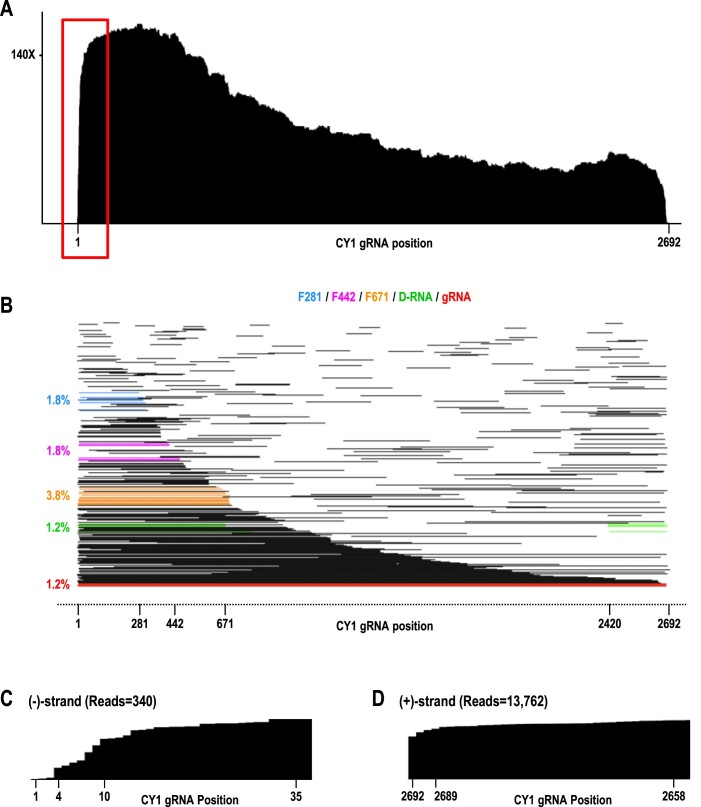
Figure 4.
Coverage map and read alignment plot of single-stranded (–)-strands in 6-wpi leaves. (A) Coverage map. (B) Read alignment plot. Reads corresponding to full-length gRNA, D-RNA, F281, F442 and F671 fragments are colored as in Figure 2, with (–)-strand read 3′end positions permitted to be somewhat less precise than for (+)-strands (allowed to be within 30 nt rather than only 10 nt of their target RNA 3′end), due to the lower precision of (–)-strand 3′end sequences. Numbering of CY1 nucleotides is for the corresponding (+)-strand. (C) Enlargement of the boxed region from A showing the missing 3′ terminal CCC in all (–)-strand reads that cover this location. (D) Comparable region from 6-wpi (+)-strand reads that covers the 3′ terminal region (3′ terminal sequence is also CCC-OH), showing no loss of reciprocal nucleotides. X-axis was flipped to aid in comparison with (–)-strand reads.