FIGURE 2.
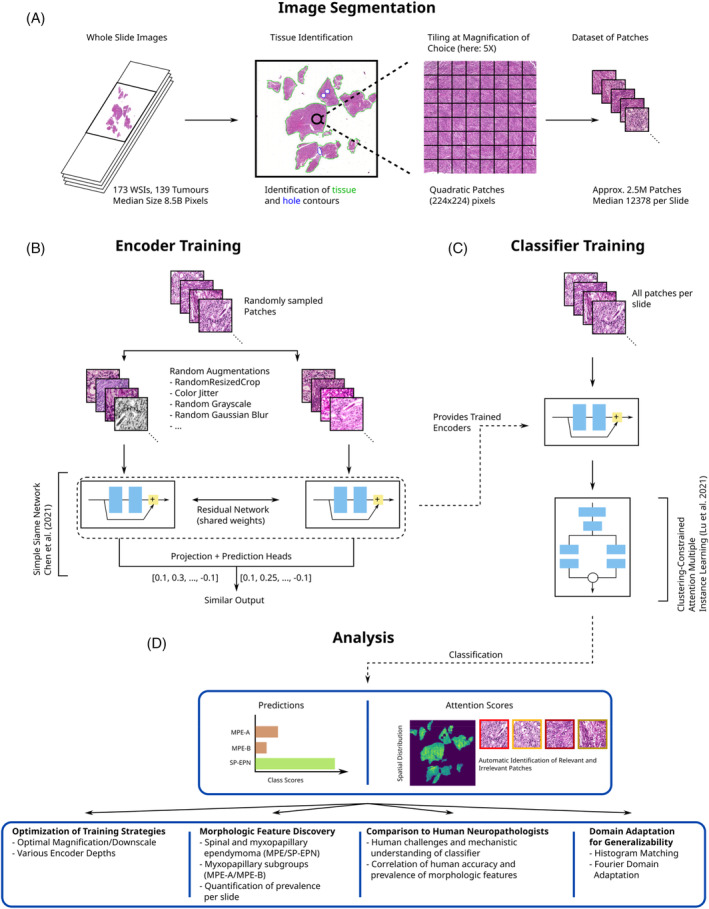
Method for the morphology‐based molecular classification of spinal cord ependymomas using deep neural networks. (A) Slide background and tissue holes are identified for each whole‐slide image (WSI) and patches are sampled from a regular grid over the tissue area. (B) Simple siamese networks are used to train residual networks on patches of WSIs from the training set. (C) The trained encoders are used for slide‐level classification using clustering‐constrained attention multiple instance learning (CLAM). (D) CLAM provides the molecular type prediction per slide, as well as the attention score per patch, for further analyses.
