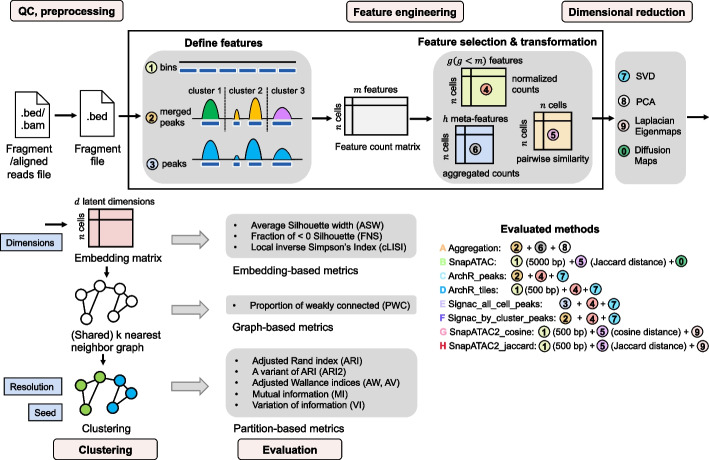
Fig. 1.
Benchmark framework. Starting from the .bed format fragment files or the .bam format aligned reads files, barcode-level QC is performed, and the filtered fragment files are input to the benchmark pipeline. The feature engineering step consists of two stages: (i) defining the genomic features and (ii) feature selection and/or transformation. Approaches for each stage are listed, and respective approaches of each method are indicated. Next, dimensional reduction is performed to generate the cell embedding matrix. A shared nearest neighbor (SNN) graph is constructed from the embedding matrix and then used for Leiden clustering. Evaluations are conducted on the embedding matrix, SNN graph, and final partitions, each using different metrics. During the evaluation, we explored multiple values for parameters such as the number of latent dimensions, resolution, and random seed―their positions within the workflow are denoted by blue boxes