Abstract
At least 20 million hepatitis E virus (HEV) infections occur annually, with >3 million symptomatic cases and ~60,000 fatalities. Hepatitis E is generally self-limiting with a case fatality rate of 0.5–3% in young adults. However it can cause up to 30% mortality in pregnant women in the third trimester, and can become chronic in immunocompromised individuals such as those receiving organ transplants or chemotherapy and individuals with HIV infection. HEV is transmitted primarily via the faecal–oral route, and was previously thought to be a public health concern only in developing countries. It is now also being frequently reported in industrialized countries, where it is transmitted zoonotically, or through organ transplantation or blood transfusions. Although a vaccine for HEV has been developed, it is only licensed in China. Additionally, no effective, non-teratogenic and specific treatments against HEV infections are currently available. Although progress has been made in characterizing HEV biology, the scarcity of adequate experimental platforms has hampered further research. In this review, we focus on providing an update on the HEV lifecycle. We will further discuss existing cell culture and animal models and highlight platforms that have proven to be useful and/or are emerging for studying other hepatotropic (viral) pathogens.
In the late 1970s, a large-scale waterborne epidemic of hepatitis spread through 200 villages in the Kashmir Valley of India, causing 52,000 cases of icteric disease and 1,700 deaths1. Although the patients’ clinical symptoms resembled hepatitis A, they were seronegative for both hepatitis A virus (HAV) and HBV. This finding led to the proposed existence of a new “enteric non-A non-B hepatitis” (ENANBH)2,3. A few years later in 1983, similar symptoms were noticed in an outbreak at a Soviet military camp in Afghanistan. A volunteer ingested pooled stool extracts from nine affected patients, and developed the typical signs and symptoms of acute hepatitis. This finding established that the virus could be transmitted via the faecal–oral route and led to the identification of 27–30 nm spherical virus-like particles in the patient’s stool that produced hepatitis when inoculated in cynomolgus monkeys4. The first partial cDNA of ENANBH was cloned and sequenced in 1990, and ENANBH was renamed ‘hepatitis E virus’ (HEV) the same year5. HEV, an RNA virus, is now recognized as a global health problem in both developing and industrialized regions including South and East Asia, East Africa, Mexico, Western Europe and the USA6–24. In this Review, we provide a summary of current knowledge on HEV, highlight cell culture and animal models that have advanced our understanding of the virus, and discuss areas in which the existing models can be improved.
Epidemiology
The global burden of hepatitis E is high; every year there are an estimated 20 million events of HEV infection, 3.3 million symptomatic cases, and 60,000 deaths attributed to HEV genotypes 1 and 224,25. Genotypes 1 and 2 are limited to humans and mostly affect developing countries, where the virus is transmitted through faecally contaminated drinking water1. HEV accounts for 50% of acute hepatitis cases in India, and has caused 17 reported large-scale epidemics in Africa between 1988 and 2013 18–20,26–39. Although large outbreaks of HEV are limited to developing countries, an increasing number of autochthonous cases are being identified in the developed world, where the prevalent HEV strains are genotypes 3 and 440. In developed nations, the primary routes of HEV transmission are zoonotic (for example, consumption of undercooked pork) and blood transfusions or organ transplants from infected donors 41–44. HEV is estimated to have a 6% seroprevalence rate in the USA, with higher prevalence in many European countries: for example, in southern France, HEV seroprevalence is 39.1% among blood donors on average, but ranges 21.9–71.3% depending on the geographical area40,45. HEV prevalence is probably underestimated as many practitioners do not routinely test for HEV in the presence of acute hepatitis symptoms, and seroprevalence studies have used serological assays with low sensitivity46,47. The assays used to detect anti-HEV IgG concentrations in serum or plasma vary considerably in sensitivity and are not standardized, complicating the interpretation of available seroprevalence data48. In Europe, awareness of HEV has been increasing over the past 10 years – studies by bloodbank centers in Denmark, France, Germany, Ireland, Netherlands, Spain, and the UK have found that 0.02–0.14% of blood donations are positive for HEV RNA49–54. Currently, blood transfusions are routinely screened for HEV RNA in Ireland and the UK, and the Netherlands have started screening blood transfusions in 201755. Selective screening occurs in France and Germany for high-risk patients, and blood authorities in Greece, Portugal, Spain, and Italy are currently evaluating whether to implement HEV screening55. In the United States, only 0.002% of plasma donations were shown to be positive for HEV RNA, suggesting that screening plasma-derived products in the US may not be necessary given the poor utility and low number of donors with positive for HEV RNA56.
HEV classification and transmission
The HEV strains affecting humans are classified into genotypes 1, 2, 3, 4, and most recently 7, and fall under the species Orthohepevirus A (Figure 1) 57. Outside of Orthohepevirus A, there are several species of HEV that infect animals but are not transmissible to humans: Orthohepevirus B (chicken), Orthohepevirus C (rat, ferret), Orthohepevirus D (bat), and the genus Piscihepevirus A (trout)57. HEV genotypes 1 and 2 of Orthohepevirus A are restricted to humans, primarily water-borne and associated with epidemics and sporadic cases in developing countries. Infections with HEV genotypes 1 and 2 are generally self-limiting and not associated with chronic disease, and are endemic to resource-poor regions in many countries in Asia, Africa and Latin America58. By contrast, HEV genotypes 3, 4 and 7 of Orthohepevirus A are primarily zoonotically transmitted through the consumption of animal products, and are associated with sporadic or cluster cases of hepatitis in industrialized countries59. Chronic cases of hepatitis E caused by infections with HEV genotypes 3, 4 and 7 have been reported in immunocompromised individuals, such as organ transplant recipients and individuals infected with HIV40,60. Although HEV infects a broad range of species including bats, ferrets, rabbits and chicken, the primary species that are considered reservoirs for transmission to humans are swine, deer and wild boar61. Of these, swine are arguably the biggest reservoir of infection and mostly likely source of zoonotic infections, with HEV RNA detected in 73% of swine farms in Sweden (based on measurements of swine faeces), 47% of swine herds in Spain (based on detection of HEV RNA in swine sera), and 24% of pig farms in France (based on presence of HEV RNA in swine liver)62–64. Infection in swine is subclinical, causing only mild hepatic lesions, and therefore swine are not routinely tested for HEV infection65. Studies measuring the presence of HEV RNA in commercial pork-based food products detected genotype 3 RNA in 47% of pork pâtés (Canada), 22% of pork liver sausages (Germany), and 30% of figatelli (French/Corsican liver sausage) samples (France)66–68. HEV can be inactivated by heating at 71°C for 20 min, therefore transmission primarily occurs through the consumption of undercooked food products69. HEV genotypes 7 and 8 infect dromedary and Bactrian camels, and there is some limited evidence of genotype 7 transmission to humans from the consumption of camel milk and meat70–72. Additional zoonotic hosts have been reported, including moose, rat, ferret, wild boar and dolphin, where it is unknown whether the corresponding HEV strains are transmissible to humans57. Little is known about the mechanisms underlying the host ranges of the various HEV genotypes.
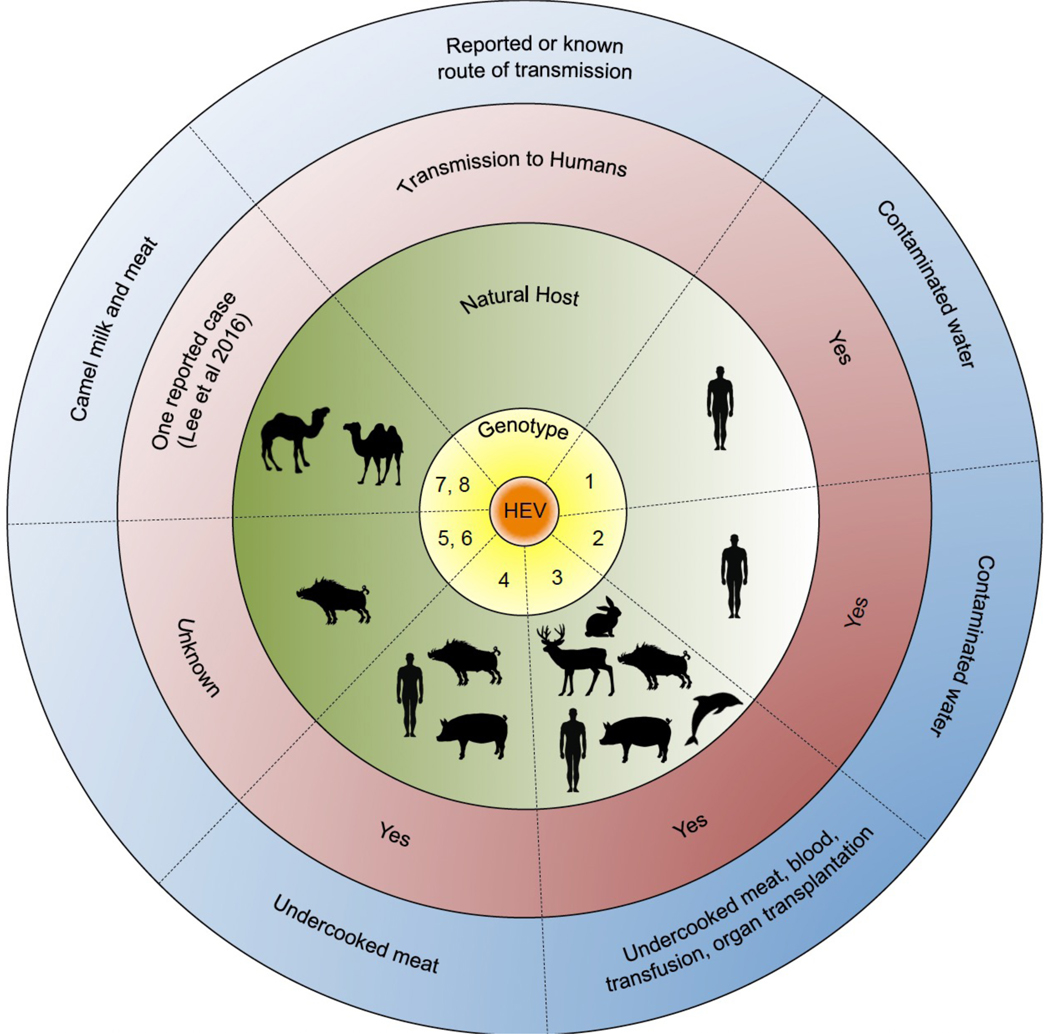
Fig 1. Host range of hepatitis E virus.
The Orthohepevirus A genus is classified into hepatitis E virus (HEV) genotype 1–8. Genotypes 1 and 2 are limited to human hosts and are transmitted via the faecal–oral route, primarily through contaminated water. Genotypes 3 and 4 have multiple hosts, and can be transmitted to humans through the consumption of undercooked meats, including pork. Genotypes 5 and 6 are known to infect wild boar; however, it is unknown whether these genotypes can be transmitted to humans (although there have been reports of wild boar genotype 3 HEV transmission to humans). Finally, genotypes 7 and 8 infect dromedary and bactrian camels, respectively. There has been one case reported of genotype 7 HEV transmission to a liver transplant patient who consumed camel meat and milk.
Clinical manifestations
Hepatitis E most commonly manifests as self-limited acute hepatitis, causing symptoms of anorexia, nausea, vomiting, malaise, abdominal pain and jaundice typically lasting ≤1 month. HEV infection is clinically indistinguishable from HAV infection, and is associated with a 1–2% mortality in immunocompetent patients24. A poorly understood clinical consequence of HEV is its severe effect in pregnant women, in which HEV infection can cause acute liver failure, haemorrhage and stillbirth, and result in up to 25% mortality in the third trimester73–75. The mechanisms underlying increased HEV virulence in pregnant women are unknown, but could be related to hormonal and/or immunological changes during pregnancy75. Acute infection has also been associated with high mortality among children under 2 years of age, and is more severe among patients with pre-existing liver disease39,76. HEV genotypes 3, 4 and 7 can become chronic in immunocompromised patients, such as organ transplant recipients and individuals infected with HIV73,77–79. These patients are at higher risk of developing chronic infection and rapid progression to cirrhosis60,80,81. Notably, evidence has emerged that commonly used immunosuppressive drugs such as tacrolimus can increase the risk of developing chronic HEV in solid organ transplant (SOT) recipients82. For example, it has been reported that the odds ratio of developing chronic HEV when using tacrolimus is 1.87 (CI: 1.49–1.97, p<0.004) as compared to when using cyclosporine A for immunosuppression82. Additionally, SOT recipients who were seropositive at the time of transplantation can become reinfected upon taking immunosuppressive therapy, and the infection can progress to chronicity83. A recent case study reported on a patient presenting with primary hepatocellular carcinoma (HCC) who was positive for HEV but not other chronic hepatitis viruses including HBV and HCV, which are commonly associated with HCC84. While these data do not prove any causal relationship, they may warrant further analysis on whether persistent HEV can culminate in HCC.
HEV primarily replicates in the liver but it has been associated with a number of extrahepatic symptoms. Correlations have been found between HEV and pancreatitis, neurological symptoms (most commonly neuralgic amyotrophy and Guillain–Barré syndrome), haematological disorders (including severe thrombocytopenia), glomerulonephritis and mixed cryoglobulinaemia, and cutaneous T cell lymphoproliferative disorders85–90. Direct causation between HEV and neurological symptoms remains to be proven, but 12 cases have been reported in which HEV RNA was detected in the cerebrospinal fluid of patients demonstrating neurological symptoms such as Guillain-Barre syndrome and neuralgic amyotrophy, and HEV has additionally been shown to replicate in neurons in vitro79,88,91,92. HEV has also been shown to replicate in intestine, lymph nodes, spleen and kidney in a swine model93. These findings reveal that hepatitis E is a complex disease whose pathogenesis and clinical progression needs to be characterized more thoroughly.
Treatment
There is currently no direct-acting treatment for HEV infection, and it remains a major public health concern particularly among immunocompromised patients and pregnant women. If possible, in transplant recipients or other immunocompromised patients, reduction of immune suppression is attempted first, which results in a sustained virologic response in 30% of patients (defined as undetectable HEV RNA in serum for 4 weeks)60,82,94. The current treatment of choice for HEV infection in chronically infected patients is monotherapy with the nucleoside analogue ribavirin; however, ribavirin is specifically contraindicated in pregnant women, who disproportionately have adverse effects as a result of HEV infection95. No established treatment for HEV is available for pregnant women, so only supportive care is provided (also known as symptomatic treatment), resulting in up to 30% maternal mortality associated with fulminant hepatic failure, spontaneous abortion and stillbirth96. Additionally, ribavirin-resistant HEV strains; for example, a genotype 3 HEV strain with a mutation in the C-terminal of the viral polymerase (encoding the G1634A protein variant) are being reported with increasing frequency in nonpregnant, chronically infected patients97. Some mutations that have been associated with ribavirin treatment failure clinically – such as the G1634R and Y1320H variants – have not resulted in ribavirin sensitivity in vitro, but have led to increased viral replication97,98. Alternative treatments for ribavirin failures, which include pegylated interferon, have met with limited success and have not been systematically evaluated99–101. Finally, preliminary studies suggest that sofosbuvir, an HCV-specific direct-acting antiviral agent, inhibits replication of genotype 3 HEV in vitro; however, this finding remains to be independently and clinically verified102,103. Ultimately, there is a great need for novel therapies against HEV97,98,104,105.
Prevention efforts have focused on sanitation, as the primary route of HEV transmission worldwide is contaminated water, and on vaccination. Hecolin (Innovax, China), a protein-based HEV vaccine eliciting anti-capsid antibodies and inducing a vigorous T cell response, is the only option for vaccination but is currently only licensed in China106–108. A second protein-based vaccine, rHEV, also contains amino acids from the capsid protein (ORF2) and was tested in phase II clinical trials but despite good safety and efficacy profile was not further developed due to the cost of clinical trials and development 109,110. Vaccine efficacy for Hecolin was shown to be 100% against genotype 1 HEV after three doses in a phase III trial in China, and cross-protects against genotype 4111. Notably, Hecolin has not been tested in pregnant women, but immunogenicity has been confirmed in pregnant mice111,112. More studies are needed to confirm whether Hecolin is effective against other genotypes including genotype 3, and to evaluate its efficacy in pregnant women and also to test efficacy in other patient populations beyond China.
An HEV vaccine holds great promise for preventing disease among residents and travellers to endemic regions such as Southeast Asia, and for reducing the alarming HEV-associated mortality in pregnant women. However, the vaccine will need to be made available outside of China, a move that is hindered by HEV not being on the WHO’s prequalification vaccine priority list113. Furthermore, distribution of the vaccine in high-endemicity regions, where health care access is often limited, will be an obstacle. Still, the vaccine holds potential to benefit high-risk patients in developed nations as well, such as SOT recipients who could be administered the vaccine as a preventative measure. Ultimately, widespread distribution of the HEV vaccine will most likely depend on public and private sector partnerships.
The molecular virology of HEV
The HEV virion is icosahedral in shape and measures 27–32nm in diameter. The capsid consists of a single, self-assembling protein whose crystal structure has previously been elucidated114. HEV was declared to be a ‘quasi-enveloped’ virus in 2016, existing in both non-enveloped and enveloped (‘eHEV’) forms, similar to HAV115. HEV is shed in faeces as a non-enveloped virus, but HEV produced in cell culture contains a lipid envelope116. The quasi-enveloped nature of HEV affords protection from neutralizing antibodies against the ORF2 and ORF3 proteins in the ‘eHEV’ form; however, attachment and entry of eHEV particles is far less efficient than that of non-enveloped HEV particles117–119. Non-enveloped HEV and eHEV are believed to have distinct cellular entry mechanisms, and further studies are required to characterize these processes117.
The HEV virion contains a positive-sense, single-stranded RNA genome of ~7.2kB in length, and is classified in the Hepeviridae family. The viral genome is organized into three open reading frames (ORFs 1–3), and contains three short untranslated regions (UTRs)120,121. A fourth open reading frame (ORF4) has been described only in HEV genotype 1, and is translated into a protein that enhances activity of the RNA-dependent RNA polymerase (RdRp)122 (Figure 2). ORF1 is the largest viral gene product of HEV, and encodes the non-structural proteins of the virus including the RdRp, RNA helicase and methyltransferase123–125. HEV also contains several other, less well-characterized domains including the ‘X’ and ‘Y’ domains, the hypervariable region (HVR), and a putative papain-like cysteine protease (PCP). HEV plausibly contains a PCP based on bioinformatic comparison that identified a domain in HEV distantly related to the protease of rubella virus126. Most positive-strand RNA animal viruses, including alphaviruses, togaviruses and picornaviruses, contain a protease that mediates polyprotein processing; however, for HEV, experiments attempting to show protease function and ORF1 polyprotein processing have yielded conflicting data 127–129. The fate of the ORF1 polyprotein and the function of the PCP-like domain of HEV both remain rich areas for further research. ORF2, the second largest viral gene, is located downstream of ORF1 and encodes the viral capsid protein of HEV130. The ORF2 capsid is highly immunogenic and is the basis for the Hecolin HEV vaccine, however in the quasi-enveloped state eHEV virions are resistant to anti-ORF2117. Although no clinical data currently suggest that the presence of eHEV limits vaccine efficacy, it could have an effect on viral spread once an infection is already established. Finally, ORF3, which is only 360bp in length, almost entirely overlaps with ORF2 and encodes a functional ion channel that is critical for release of infectious viral particles131. ORF3 has additionally been shown to interact with a variety of host proteins including tumor susceptibility gene 101 protein (TSG101), a key component of the endosomal sorting complexes required for transport (ESCRT) pathway that is used by a number of viruses (including HIV) for budding of progeny virions132,133.
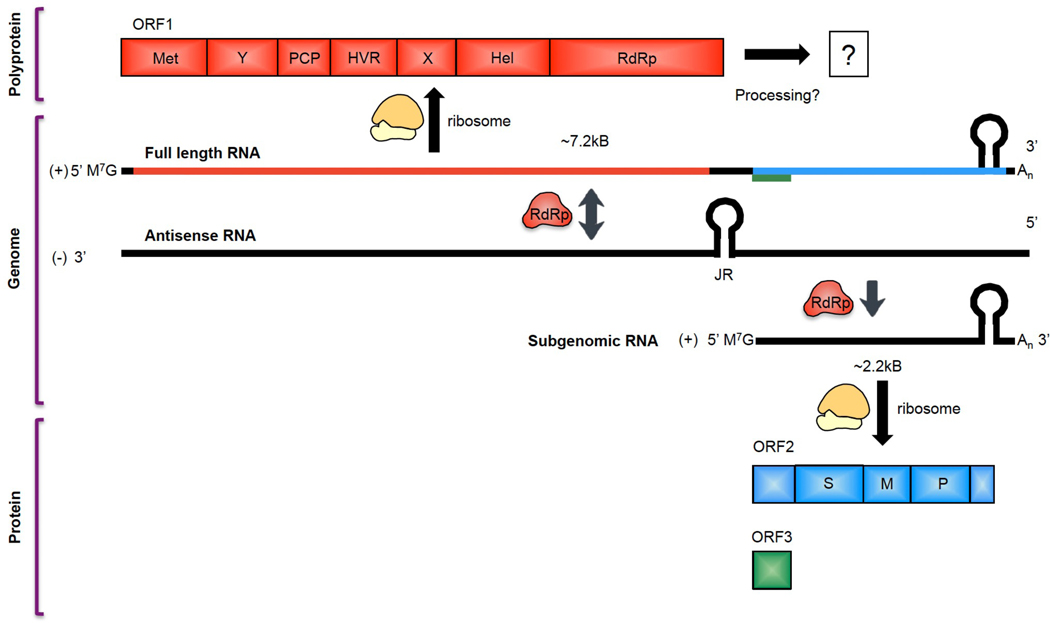
Fig 2. Genetic organization and translation of hepatitis E virus.
Hepatitis E virus (HEV) is a ~7.2kB, positive (+)-sense single-stranded RNA virus. The mRNA is capped at the 5’ end, polyadenylated at the 3’ end, and the junctional region (JR) between ORF1 and ORF2/3 contains a stem-loop structure that is critical for HEV replication. After viral entry and uncoating, the (+)-sense full-length viral genome is translated by host ribosomes to produce the ORF1 polyprotein, which contains the non-structural replication machinery of the virus including the methyltransferase (Met), RNA helicase (Hel), and RNA-dependent RNA polymerase (RdRp), as well as several non-enzymatic regions essential for efficient viral replication (the ‘Y’, ‘X’, and ‘hypervariable’ (HVR) regions). Additionally, ORF1 contains a putative papain-like cysteine protease (PCP) based on sequence similarity to the protease of rubella virus, though data showing protease activity for this region have been conflicting. It is unclear whether the ORF1 polyprotein undergoes processing into smaller units. HEV genotype 1 is thought to contain an additional open reading frame, ORF4, that is translated into a viral protein enhancing RdRp activity. After translation of the ORF1 polyprotein, the RdRp from ORF1 transcribes an antisense (–)-stranded intermediate RNA from the (+)-sense strand. The (–)-sense strand then serves as a template for the transcription of more (+)-sense full-length RNA for packaging into new progeny virions, as well as a shorter, ~2.2kB subgenomic RNA (sgRNA) encoding ORF2 and ORF3. These viral genes are ~2.2kB and ~360bp in length, respectively, and ORF3 entirely overlaps with ORF2 except for one leading base pair. The sgRNA, which is capped at the 5’ end and polyadenylated at the 3’ end, is then translated into the ORF2 capsid protein and the ORF3 viroporin based on a leaky scanning mechanism by host ribosomes. Regulation of transcription of the sgRNA is poorly understood.
As for many other viruses heparan sulfate proteoglycans (HSPG) are required for the attachment of HEV virions to their target cells, and eHEV enters cells through a process that involves receptor-dependent clathrin-mediated endocytosis, the Rab5 and Rab7 GTPases and lysosomal lipid degradation117,134–136. Non-enveloped HEV and eHEV are believed to have distinct entry mechanisms, and little is known about entry mechanisms for non-enveloped HEV specifically. A cell surface receptor mediating HEV entry also remains to be identified. Upon viral entry of eHEV, the quasi-envelope is believed to undergo lysosomal lipid degradation to expose the capsid protein117. The virion then uncoats in a poorly understood process, and the positive-sense HEV RNA is translated by host factors to produce the ORF1 polyprotein containing the RdRp. The RdRp then transcribes complementary full-length negative-sense viral RNA, which serves as a template for transcribing positive-strand full-length HEV RNA, and a 2kB subgenomic transcript encoding ORF2 and ORF3137−140 (Figure 3). Limited evidence suggests that an additional 3.7kB subgenomic RNA might be transcribed from the negative-sense template137,141,142. Both the full-length and subgenomic transcripts are capped and polyadenylated137. Host ribosomes are then thought to use leaky scanning to translate the ORF2 protein (pORF2) and ORF3 protein (pORF3) from the subgenomic RNA in the endoplasmic reticulum143. Leaky scanning refers to a phenomenon in which a ribosome will occasionally skip a ‘weaker’ initiation codon – possibly an ‘ATG’ triplet in a weak Kozak consensus sequence – and instead use a second, downstream initiation codon; in this way the ribosome can translate two proteins from the same transcript. The regulation and relative levels of translation of these proteins are not well understood. The pORF2 capsid is then processed in the endoplasmic reticulum and glycosylated at three sites, a modification necessary for the formation of infectious virions130,144,145. pORF2 binds the 5’ end of HEV, an interaction that could mediate encapsidation; however, a complete picture of the role of pORF2 in the life cycle remains to be determined146. Different studies show that pORF2 localizes to the Golgi, cytoplasm and even the nucleus, suggesting that the protein could have multiple, hitherto unknown functions147,148. pORF3, a 113 amino acid phosphoprotein, is another poorly understood HEV product that is required for release of virions and was shown to exhibit ion channel activity in a paper published in 2017131. The ORF3 ion channel does not seem to have any discernible preference for specific anions or cations, and the mechanisms underlying viroporin-mediated viral release remains to be determined. Cell lysis and the subsequent release of infectious virions can be triggered by increased membrane permeability as a result of viroporin accumulation 149 but this process does not seem to have a role in HEV egress as HEV is thought to be a non-cytolytic virus. Viroporin insertion in cellular membranes have been proposed to disrupt the chemoelectrical barrier by facilitating flux across membranes, therefore dissipating the membrane potential of internal vesicles or the plasma membrane and stimulating viral budding 150.
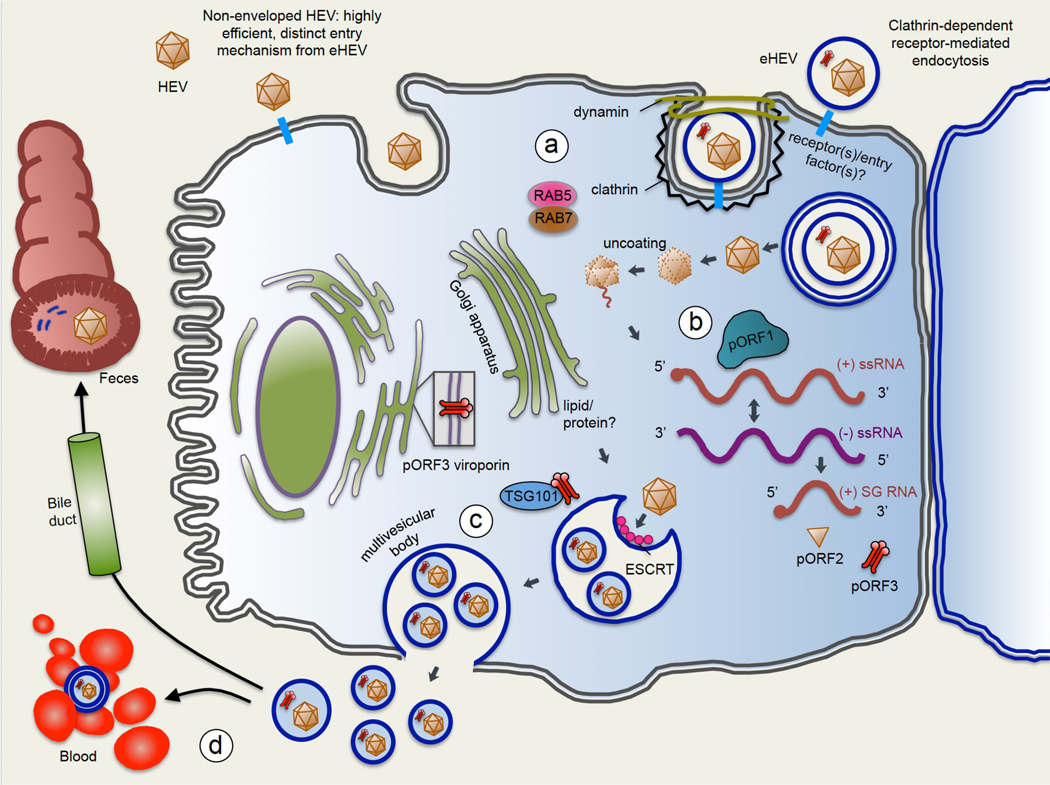
Fig 3. Life cycle of hepatitis E virus.
(1) Viral entry: hepatitis E virus (HEV) is a quasi-enveloped virus, meaning it can exist in the non-enveloped state (HEV) or can be coated in a lipid-derived membrane (eHEV). HEV and eHEV have distinct entry mechanisms. Little is known about entry mechanisms for HEV. For eHEV, the virus enters the cell through clathrin-dependent and dynamin-dependent, receptor-mediated endocytosis. A specific cell surface receptor mediating eHEV entry remains to be identified, but the GTPases Rab5 and Rab7 are known to have a role in eHEV entry. Upon entering the cell, the envelope of eHEV undergoes lysosome-mediated lipid degradation, and uncoats in a poorly understood process to expose the viral mRNA. (2) ORF1 polyprotein (pORF1) containing the RdRp is translated from the (+)-strand, and the RdRp then transcribes full-length (–)-sense RNA. The (–)-sense RNA serves as a template for transcribing more full-length (+)-sense RNA to be packaged into progeny virions, as well as a shorter subgenomic RNA (sgRNA) which encodes ORF2 and ORF3. The ORF2 capsid protein (pORF2), and the ORF3 protein (pORF3), a viroporin essential for viral release, are translated from the sgRNA. (3) pORF3 binds to TSG101, a member of the endosomal sorting complexes required for transport (ESCRT) pathway that is used by several other RNA viruses to bud from cell membranes. The interaction of pORF3 with TSG101 probably promotes budding of progeny virions into multivesicular bodies (MVBs), which then fuse with the plasma membrane to release virions from the cell. The lipid envelope of eHEV is thought to be derived from the trans-Golgi network, and viral particles contained in eHEV have been shown to be associated with pORF3. pORF3 has additionally been shown to exhibit viroporin activity, and it is possible that pORF3 exists in multiple forms to perform distinct functions. (4) eHEV released from the apical membrane enters the bile duct, where the lipid envelope is thought to be degraded by detergents and proteases in the bile. This feature would explain why HEV in the faeces is non-enveloped. On the other hand, eHEV released from the basal membrane of hepatocytes enters the serum in its quasi-enveloped form, where it is protected from neutralizing antibodies against pORF2 and pORF3, but is less efficient at infecting cells.
In addition, pORF3 interacts with a broad range of host cellular proteins including 3IP, microtubules, SH3 and Pyst1 (both leading to activation of MAPK), bikunin (serine protease inhibitor), TSG101, hemopexin, fibrinogen, HIF1A, CIN85, HNF4 and hepsin130,151–162. These diverse interactions suggest that pORF3 might modulate the host environment in multiple ways to create favourable conditions for the viral life cycle, in addition to its role in viral release163. Notably, the role of pORF3 in virion release is dependent on a highly conserved PSAP motif which enables the interaction with TSG101, a cellular factor involved in the budding of viruses and a member of ESCRT pathway132,164–171 79. The ESCRT pathway is used by a number of other RNA viruses (for example, HIV) during viral release, and involves budding of the virus through the cellular membrane, leading to the acquisition of a lipid envelope172. Thus, the ESCRT pathway would explain how eHEV particles are formed; however, this aspect suggests that non-enveloped HEV uses a different mechanism for release that is thus far unknown, or that non-enveloped HEV is formed through shedding of the lipid envelope following release. Ultimately, more studies are needed to close the numerous gaps in our understanding of the HEV life cycle.
Cell culture models
HEV has historically been extremely difficult to culture in vitro, replicating at very low titers. Early experiments developing in vitro infection systems with full-length virus used a variety of cell types including primary hepatocytes from macaques, human HepG2 hepatoma cells, A549 lung adenocarcinoma cells and simian primary kidney cells (Table 1)173–176. However, amplification was required to detect HEV in the medium of these cells, and this lack of an efficient cell culture study hampered efforts to study the HEV life cycle. Breakthroughs in developing robust in vitro systems to study HEV have been achieved not only through identifying compatible cell lines, but also through the isolation of specific strains with improved replication efficiency in vitro.
Table 1.
In vitro models to study HEV Orthohepevirus A.
| Type | Cell line | Tissue | Species | HEVgenotype | Strainstested |
|---|---|---|---|---|---|
| Immortalized | LLC-PK1 | Kidney epithelial | Swine | 3 | KernowC1/p6 |
| FRhK-4 | Kidney epithelial | Rhesusmacaque | 1 | Sar55 | |
| HepG2, HepG2/C3A | Liver hepatoma | Human | 3 | KernowC1/p6 | |
| HepaRG | Liver hepatoma | Human | 3 | KernowC1/p6 | |
| PLC/PRF/5 | Liver hepatoma | Human | 3,4 | JE03-1760F, HEJF5/15F | |
| Huh7, Huh7.5, S10-3 | Liver hepatoma | Human | 1,3 | Sar55, KernowC1/p6 (Note: S10-3 is a subclone of Huh7 cells selected for its ability to produce infectious Sar55 virus) | |
| A549 | Lung adenocarcinoma | Human | 3,4 | JE03-1760F, HEJF5/15F | |
| Caco-2 | Colon adenocarcinoma | Human | 1 | Sar55 | |
| Primary cells | Primary tissue | Liver | Hepatocyte-like | 1 | Hepatocyte-like |
| iPSC- derived hepatocyte like cells | Hepatocyte-like | Human | 3 | KernowC1/p6 | |
| Porcine embryonic stem cells | Embryonic | Porcine | 3 | Swine HEV genotype 3f |
HEV, hepatitis E virus; iPSC, induced pluripotent stem cells.
Cell lines.
Generally, genotype 1 HEV has been more difficult to culture in vitro than genotypes 3 and 4, so the available cell-culture-adapted strains are mostly derived from the latter. In 2009, the genotype 3 JE03–1760F and genotype 4 HE-JF5/15F HEV strains were isolated from infected patients and found to have increased replication efficiency in A549 cells and PLC/PRF/5 liver hepatoma cells177,178. These strains accumulated mutations after being serially passaged in cell culture, that presumably enhanced their ability to replicate in these systems. In 2011, the genotype 3 Kernow-C1 strain of HEV was isolated from a chronically infected patient who was co-infected with HIV and found to efficiently infect human, deer and pig cell lines after being serially passaged six times in culture179. The increased ability of the Kernow-C1/p6 strain to replicate in vitro was due to a 57-amino acid insertion from the human S17 ribosomal RNA into the ORF1 HVR domain. The S17-containing recombinant strain was present in the original faecal sample from the patient, who had become host to multiple quasi-species of HEV as the virus mutated during the length of his chronic infection. Over six serial passages in HepG2/C3A human hepatoma cells, the S17 insertion-containing strain was found to propagate more efficiently and become the dominant quasi-species. Introduction of the S17 insertion into a different, genotype 1 strain of HEV markedly enhanced its ability to transfect hamster BHK-21 cells180. Similarly, another genotype 3 HEV strain, LBPR-0379, was identified to contain an insertion in its HVR region from the S19 human ribosomal protein that conferred a growth advantage in cell culture181. The mechanisms whereby these insertions improve replication of HEV in vitro and broaden the virus’ host range are unknown and the subject of great interest. Furthermore, they demonstrate the ability of the virus to mutate into quasi-species and acquire novel capabilities during chronic infection.
In vitro studies of HEV have used the HepG2, HepG2/C3A, HepaRG, Huh7, Huh7.5 and S10–3 hepatoma cell lines to study viral replication118,131,182–184. HepG2/C3A, a subclone of the HepG2 hepatoma cell line, is a popular model for in vitro drug testing and exhibits an improved hepatic phenotype over HepG2 cells185. Replication of the Kernow-C1/p6 strain of HEV was reported to be ~7.5-fold higher in HepG2/C3A cells than in Huh7.5, PLC/PRF/5, and A549 cells, making the combination of the Kernow-C1/p6 strain with HepG2/C3A a powerful tool for studying HEV179. That said, hepatoma and other tumour-derived cell lines do not adequately reproduce the physiological environment of primary cells (hepatocytes) because of their abnormal cell proliferation and aberrant gene expression and regulation186.
Primary hepatocytes.
Cultures of primary hepatocytes are more desirable for in vitro experiments for HEV infection, but their use has several practical limitations. Indeed, it was shown that HEV can infect primary hepatocytes of cynomolgus monkeys, but infection of primary human hepatocytes has not been reported yet173. Once isolated, primary hepatocytes do not proliferate or undergo limited proliferation, which is a challenge. Furthermore, the phenotype of these primary hepatocytes is unstable, as they tend to de-differentiate within days in conventional culture systems, thereby precluding longer-term studies of HEV infections187. Primary human hepatocyte dedifferentiation can be delayed or prevented in collagen sandwich cultures, by aggregation in spheroids or in co-culture with non-parenchymal cells188–192. Primary human hepatocytes aggregated into spheroids have been infected with HCV193. For the latter approach, both self-assembling (SACC) and micro-patterned primary human hepatocyte co-cultures (MPCC) are effective formats to stabilize hepatic function, especially if oxidative stress is reduced during the initiation of the culture194 195,196. MPCC and SACC primary human hepatocytes have been infected with HBV, HCV and Plasmodium falciparum and P. vivax 197–200 and therefore might prove useful to establish longer-term HEV infections, possibly even with non-cell culture-adapted patient isolates. Infections in primary human hepatocytes are frequently hampered by considerable variability in the susceptibility between different hepatocyte donors although the underlying etiology of this variability is unknown.
Stem cell derived hepatocyte like cells and tissue organoids.
In 2016, induced pluripotent stem cell (iPSC)-derived hepatocyte-like cells (HLCs) were shown to support HEV replication for the Kernow-C1/p6 strain, although at lower levels than HepG2/C3A cells201. iPSCs carry the advantages of being indefinitely self-renewing, more easily amenable to genetic manipulations than fully differentiated cells, capable of differentiating into numerous cell types and able to produce patient- and tissue-specific primary — they are a promising in vitro model for studying HEV in a more physiologically relevant context than hepatoma-derived cells202–204. To better approximate the cellular complexity of the liver, efforts are under way to create more complex cultures, incorporating additional liver resident non-parenchymal cell populations including endothelial cells, cholangiocytes, Kupffer cells, fibroblasts, and stellate cells205,206. (Figure 4). Combining these cell types, preferably in physiologically relevant ratios, will be particularly helpful to more accurately mimic the inflammatory environment of the liver during HEV infection. As an additional layer of complexity, incorporating primary hepatocytes in a 3D architecture will be important to closely mimic the transcriptional heterogeneity of hepatocytes in the liver that is influenced by a variety of environmental cues, including oxygen and nutrient gradients. Differences in transcriptional activity have clearly been documented for genes involved in metabolism but could conceivably also affect the susceptibility and host response to (viral) pathogens, such as HEV207. Tremendous advances have been made in engineering very sophisticated (primary) cell culture platforms, which have already been successfully used to study other hepatotropic viruses such as HBV200,208. Undoubtedly, the HEV field will benefit from these advances and might even become a driver in refining them.
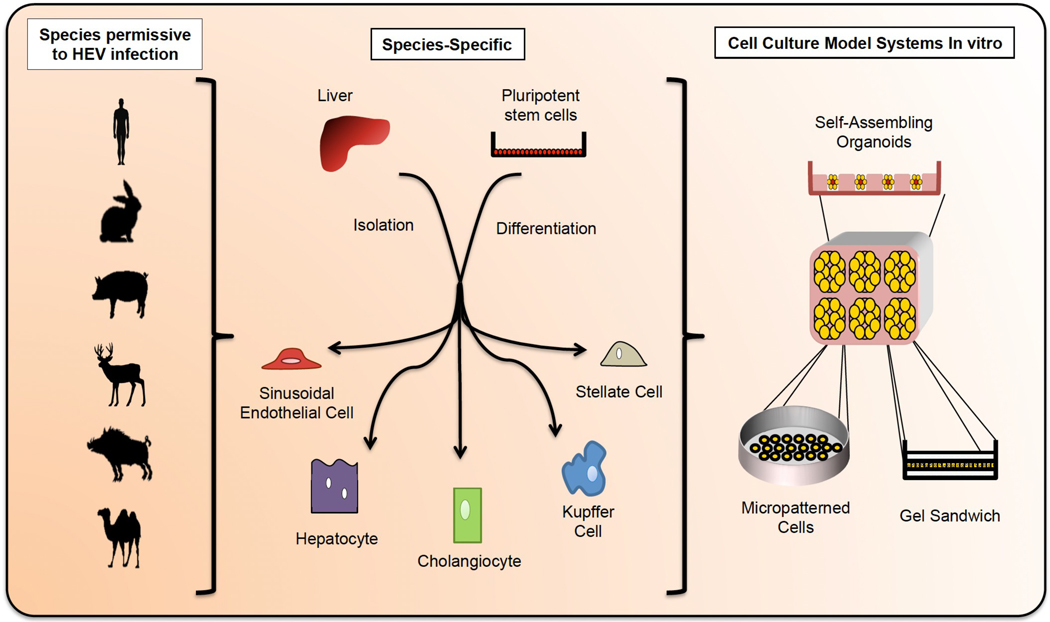
Fig 4. Towards more physiologically relevant 2D and 3D cell culture models for studying HEV.
The species permissive to HEV infection include (but are not limited to) humans, rabbits, swine, deer, wild boar, and camel. In order to better study HEV infection in physiologically relevant in vitro models, it will be desirable to generate co-cultures that recapitulate the complexity of the liver including endothelial, stellate, cholangiocytic, Kupffer, and hepatic cells in the appropriate ratios. These cells can be harvested from primary tissue or differentiated from stem cells, and could be derived from the aforementioned species to explore viral host tropism. Primary cultures have the disadvantage of limited durability; this issue can be overcome by differentiating the various cell types from indefinitely self-renewing stem cells. The latter, however, is technically challenging and requires advances in current hepatic differentiation protocols. Incorporating primary and stem-cell derived tissues into a 3D architecture will also be important to more closely mimic the physiological hepatic environment and preserve cell morphology. The architecture of the liver leads to heterogeneous environmental cues (e.g. nutrients, oxygen, inflammatory factors, etc) reaching individual cells, and 3D cultures can better capture this phenomenon. Furthermore it was previously shown that primary human hepatocyte dedifferentiation can be delayed or prevented in collagen sandwich cultures, by aggregation in spheroids, or in co-culture with non-parenchymal cells.
Tissue tropism.
It is important to note that in contrast to HBV, HCV and hepatitis delta virus (HDV), which are thought to productively infect only hepatocytes in vivo, HEV has a broader tissue tropism. HEV RNA and/or antigens have been detected in small intestine, colon, lymph nodes, placenta, dermal microvascular endothelial cells, and neurons based on studies in swine and humans90,209–212. Using negative-strand-specific PCR, HEV replication has been detected in human placenta209. This finding raises the possibility that non liver-derived cells could have potential as in vitro systems to study HEV. Indeed, HEV is capable of replicating in a number of non-liver cell types. Notably, the lung adenocarcinoma-derived A549 cell line was one of the earliest cell lines used to culture HEV, and is permissive to the genotype 3 JE03–1760F and genotype 4 HE-JF5/15F HEV strains, as well as swine, wild boar and rabbit-derived strains of HEV 177,178,213–215. HEV has also been shown to infect porcine embryonic stem cell-derived hepatic cells184. It should be clearly noted, though, that the ability of a given virus, such as HEV, to infect and replicate in a cell line, does not necessarily imply that this cell type would be a natural reservoir for the virus. Exemplary for this point is that despite replicating in A549 cells in vitro, HEV has not been correlated with pulmonary HEV-related symptoms in patients.
Animal models
Even with these advancements, tissue cultures have limited utility in studying virally induced immune responses and disease. Thus, creating suitable animal models for HEV will remain a priority to study the pathogenesis of HEV, for example, during both fulminant acute and chronic hepatitis E, and to test novel antiviral therapeutics. The optimal model should be fully immunocompetent, susceptible to genetically diverse HEV strains causing disease in humans and recapitulating clinically apparent disease. From a practical perspective, the model should be cheap, easy to propagate, amenable to genetic manipulations and optimally a plethora of reagents should be available to monitor host responses to the infection. Such a model does not exist – yet.
To bridge this gap, (at least) three alternative and certainly not necessarily mutually exclusive approaches could be taken: conceivably, surrogate models, that is species which naturally support HEV infection to some extent, could be used; the host environment of a usually resistant species could be engineered to render it more conducive to HEV infection; HEV could possibly be adapted genetically to enable the virus to overcome species barriers.
Primates.
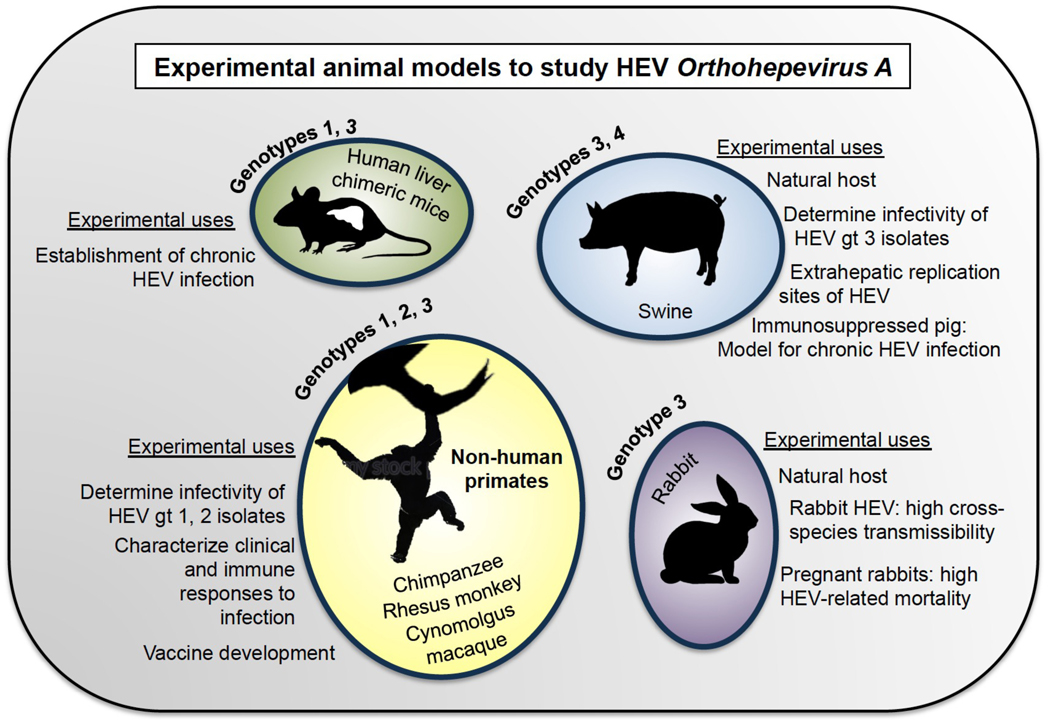
In contrast to the other hepatitis viruses, which exhibit a narrow host range largely limited to humans and closely related great apes (HBV, HCV, HDV) or at least smaller primates (HAV), certain HEV genotypes have been found in a variety of species. These include swine, deer, and rabbit that serve as reservoirs for HEV in industrialized nations 59 216. Additional zoonotic hosts include moose, rat, wild boar (genotype 5,6), camel (genotype 7,8), and dolphin (genotype 3), but it is unknown whether the corresponding HEV strains are transmissible to humans 57. Despite the broad host range of HEV, infection is subclinical in most zoonotic hosts, and tractable small animal models to study the virus in vivo are lacking. Non-human primates have been among the few animals successfully used to study genotypes 1 and 2 of HEV, in particular rhesus monkeys, cynomolgus macaques and chimpanzees (Figure 5)217–219. Numerous experiments have been done in chimpanzees using genotype 1 HEV, including analyses of transcriptomic changes associated with infection, measurement of the duration of faecal shedding and viraemia, and the discovery that capping of HEV is required for infectivity220–223. Chimpanzees and rhesus monkeys have additionally been used to show that swine HEV can cross the species barrier and infect non-human primates224. Infected primates develop clinical responses to HEV that mimic some features of human disease, including focal hepatocyte necrosis with accumulations of macrophages and activated Kupffer cells, and the anti-HEV immune response in primates is similar to that observed in humans 221,225. Thus, nonhuman primates have been useful models for studying hepatitis E clinical progression and for immunological studies220,226–228. Both cynomolgus and rhesus monkeys have been used to test the efficacy of ORF2 capsid protein-derived anti-HEV vaccines229,230 231. However, there are limitations to the use of non-human primates in modelling HEV in vivo. Attempts to reproduce HEV-associated mortality in the context of pregnancy were unsuccessful in pregnant rhesus monkeys, who also do not exhibit vertical transmission of the virus232 233. Furthermore, primates are expensive to maintain, their use raises ethical concerns, and hepatitis E infection in primates does not accurately mimic some aspects of disease progression in humans, in particular HEV-associated liver injury in the context of pregnancy232,233.
Fig 5. Experimental animal models to study HEV Orthohepevirus A.
Experimental animal models that have been used to study HEV include non-human primates, swine, rabbits, and human liver chimeric mice. Chimpanzees, rhesus monkeys, and Cynomolgus macaques were the earliest animal models in HEV research, and have been used to study HEV pathogenesis and vaccine efficacy. Swine, which are naturally infected with gts 3 and 4 of HEV and can transmit these strains to humans, have been used to determine the infectivity of gt 3 isolates, and to show extrahepatic replication sites of HEV. Recently, an iatrogenically immunosuppressed swine model was shown to support chronic infection with gt 3, and similarly, human liver chimeric mice can support chronic infection with gt1 and gt3 HEV. Gt, genotype.
Swine.
Swine were discovered to be natural hosts to HEV in 1995, and are now known to be the primary route of HEV transmission to humans in developed countries234,235. Unlike rhesus monkeys, which can be infected with HEV genotypes 1–3, swine are only susceptible to genotypes 3 and 4236,237. Infection in swine is largely subclinical, but pigs develop relatively more severe hepatic lesions when infected with human genotype 3 HEV than swine genotype 3 HEV65,235,238–241. Swine have been used to demonstrate extrahepatic sites of HEV replication, including small intestine, lymph nodes and colon, and for studies on cross-species infection211,242. In 2017, experimental pigs treated with immunosuppressive drugs were used to successfully establish chronic HEV infection with genotype 3, the same genotype responsible for chronic infection in humans243. Thus, swine have proven to be useful models for studying genotypes 3 and 4 of HEV, but are not susceptible to genotype 1, which accounts for the majority of clinical cases in humans worldwide.
Rabbits.
Rabbits are another natural host of HEV who could serve as useful models for HEV studies244. Rabbit strains of HEV have been experimentally shown to infect swine and cynomolgus macaques, demonstrating a high potential for cross-species transmission245,246. Rabbits show limited clinical symptoms from HEV, but notably studies from one group suggest that rabbits support chronic infection and extrahepatic replication of HEV, and that pregnant rabbits have high HEV-associated mortality247,248. Given the relatively small size of rabbits and the potential for transmission of rabbit HEV strains to humans, rabbits might be an interesting model in which to explore the pathogenesis of hepatitis E.
Small animals.
There has been limited success infecting naive mice with HEV, who are not natural hosts for the virus. One study reported that Balb/c nude mice were susceptible to genotype 4 HEV isolated from swine, however these findings have not been confirmed independently249. Conceivably, mouse orthologues of certain yet-to-be-identified host factors only inefficiently support different aspects of the viral life-cycle. Greater understanding of the different aspects of the viral life-cycle and essential cellularly encoded co-factors would potentially allow us to overcome the species barrier of HEV genetically and to create mouse models that robustly support HEV infection.
Alternatively, tissue humanization approaches have been effective in establishing infections with other human hepatotropic pathogens. Mice growing a partially human liver can support infections with hepatotropic pathogens in vivo, including HBV, HCV, HDV and liver stages of parasites causing malaria in humans250–260. Humanized mice are usually generated through transplantation of human hepatocytes into immunocompromised liver injury recipients. The resulting human liver chimeric mice have been used to successfully establish infection with HEV genotypes 1 and 3 in the past few years261–264. These xenotransplanted mice are a tractable and valuable model for drug testing and for studying long-term viral persistence within the 3D context of the liver. However, a considerable shortcoming of singly engrafted human liver chimeric mouse models is their inability to mount cellular and humoral immune responses due to their highly immunocompromised status that is necessary to prevent graft rejection. To study HEV-specific immune responses, which counteract the infection but are also thought to contribute to the progression of liver pathogenesis, xenorecipients co-engrafted with both human hepatocytes and components of a human immune system in a single recipient might prove useful. Such dually engrafted mice have been used in to study human immune responses to HBV and HCV for example265–267. However, given that the human immune response is generally weak in such humanized mouse models, continued refinements of the xenorecipients strains and engraftment procedures remain critical.
Rats are another potential rodent model for HEV, and unlike mice, are natural hosts for HEV268. However, the HEV strains infecting rats are classified under Orthohepevirus C, and are only distantly related to the human-tropic strains of HEV, which are classified in Orthohepevirus A. One group successfully infected athymic nude rats with rat HEV, however rats are not susceptible genotypes 1, 2, or 3 of human HEV, and furthermore rhesus monkeys are not susceptible to rat HEV269,270. Although it is conceivable to generate HEV chimeras between strains of Orthohepeviruses A and C, this approach could prove difficult because of genetic incompatibilities that could compromise the fitness of these genomes. Finally, gerbils are another rodent model that warrant further investigation for their potential as a tool to study HEV. Several groups have reported successful infection of Mongolian gerbils with genotype 4 HEV271–273.
Conclusions
Although HEV is becoming increasingly recognized, much work remains to be done in understanding its pathogenesis and molecular mechanisms (Box 1). Little is known about key aspects of the viral life cycle – for example, the cellular (co-)factors involved in different steps in the viral life-cycle, the most prominent receptor(s) mediating viral entry, whether and how polyprotein processing occurs for ORF1, and the role of ORF3 in viral release. A better understanding of these mechanisms could hold the key to developing direct-acting antiviral therapeutics. The clinical pathogenesis of hepatitis E disease contains many mysteries as well, from the high mortality rate in pregnant women caused by specific strains of the virus, to the many extrahepatic symptoms that are being reported in association with infection. Ultimately, more clinical data and better cell culture and animal models are needed to understand interactions between the virus and its host. Finally, as we encroach into new ecological spaces, more accurate epidemiological data is needed to understand the transmissibility of HEV from foods, and to characterize the full extent of the host range of HEV.
Box 1 |. Key questions or challenges in HEV research.
Gaining a thorough mechanistic understanding of crucial aspects of the viral lifecycle: entry, genome replication, assembly and release
Biophysical analysis and biochemical composition of hepatitis E virus (HEV) particles
(noneveloped HEV versus enveloped HEV (eHEV))
Obtaining high-resolution structures of pORF1 and pORF3
Identification of essential host factors governing different aspects of the HEV lifecycle
Defining the HEV host and tissue tropism
Mechanism of HEV-mediated pathogenesis in clinically relevant settings (e.g. acute liver failure during pregnancy and extrahepatic manifestations)
Effect of HEV co-infection in patients with underlying liver disease, such as chronic viral hepatitis
Creating robust cell culture models supporting infection with all HEV genotypes
Developing tractable (small) animal models that adequately recapitulate disease symptoms observed in patients
Collecting epidemiological data on the prevalence and transmissibility of different HEV genotypes in humans and animals
Mechanisms of innate immune recognition and correlates of immunological protection
Developing direct-acting or host-targeting antiviral agents that can effectively cure HEV infection and can be administered to all patient populations
Key points.
HEV causes varying disease severity among patient subpopulations: it is self-limiting in most young adults, but causes ~30% mortality in pregnant women, and lead to chronicity in immunocompromised patients.
HEV has a broad but poorly characterized host range, and in industrialized countries it is primarily transmitted zoonotically through the consumption of undercooked meat.
A prophylactic vaccine against HEV exists but is currently only licensed in China.
There is currently no direct-acting therapy available against HEV, and no non-teratogenic treatment options for pregnant women, creating a need for development of new therapeutics.
The molecular biology of HEV remains incompletely understood.
New model systems are emerging to study HEV, but more refined models are needed to gain insights in the interactions of HEV with its host including mechanisms of HEV pathogenesis.
Acknowledgements:
We thank members of the Ploss lab for critical discussions of the manuscript. The work was supported in part by grants from Princeton University and an Investigator in Pathogenesis Award by the Burroughs Wellcome Fund (to A.P.). Q.D. is supported by a postdoctoral fellowship from the New Jersey Commission for Cancer Research. We apologize to all authors whose work could not be cited due to space constraints.
Author biographies:
Ila Nimgaonkar is an MD/PhD student in the Rutgers-Robert Wood Johnson Medical School-Princeton University physician-scientist training program. She is interested in viral factors governing host tropism in hepatitis E virus, and in understanding basic mechanisms of the HEV life cycle.
Qiang Ding is a senior research associate in the Department of Molecular Biology at Princeton University. His current research focuses on study mechanisms of HEV replication and assembly. He is also interested in host range restrictions of acute and chronic hepatotropic human viral pathogens.
Robert Schwartz is an Assistant Professor in the Departments of Medicine and Physiology, Biophysics and Systems Biology at Weill Cornell Medicine. His research focuses on the derivation of stem cell derived hepatocytes, primary human hepatocytes and other stem cell derived lineages and on leveraging their use as models to study metabolic and infectious disease.
Alexander Ploss is an Assistant Professor in the Department of Molecular Biology at Princeton University where he is also a Faculty Affiliate in the Program in Global Health and Health Policy. He is also a full member of the Cancer Institute of New Jersey. His research focuses on analyzing the host tropism of human hepatotropic pathogens and the construction of cell culture and humanized animal models to study host responses to infection.
Footnotes
Competing interests statement
A.P. and Q.D. are inventors on a patent application defining an hepatitis E virus transcomplementation system for antiviral drug screening and ORF3’s viroporin function as a antiviral drug target.
References
- 1.Khuroo MS, Khuroo MS & Khuroo NS Hepatitis E: Discovery, global impact, control and cure. World J Gastroenterol 22, 7030–7045, doi: 10.3748/wjg.v22.i31.7030 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khuroo MS Study of an epidemic of non-A, non-B hepatitis. Possibility of another human hepatitis virus distinct from post-transfusion non-A, non-B type. Am J Med 68, 818–824 (1980). [DOI] [PubMed] [Google Scholar]
- 3.Wong DC, Purcell RH, Sreenivasan MA, Prasad SR & Pavri KM Epidemic and endemic hepatitis in India: evidence for a non-A, non-B hepatitis virus aetiology. Lancet 2, 876–879 (1980). [DOI] [PubMed] [Google Scholar]
- 4. Balayan MS et al. Evidence for a virus in non-A, non-B hepatitis transmitted via the fecal-oral route. Intervirology 20, 23–31 (1983). A volunteer ingested pooled stool extracts from patients infected with HEV to show for the first time that the virus is fecal-orally transmitted.
- 5. Reyes GR et al. Isolation of a cDNA from the virus responsible for enterically transmitted nonA, non-B hepatitis. Science 247, 1335–1339 (1990). The first partial cDNA of ET-NANBH was cloned, and the virus renamed ‘hepatitis E virus’.
- 6.Arankalle VA, Chadha MS, Mehendale SM & Banerjee K. Outbreak of enterically transmitted non-A, non-B hepatitis among schoolchildren. Lancet 2, 1199–1200 (1988). [DOI] [PubMed] [Google Scholar]
- 7.Naik SR, Aggarwal R, Salunke PN & Mehrotra NN A large waterborne viral hepatitis E epidemic in Kanpur, India. Bull World Health Organ 70, 597–604 (1992). [PMC free article] [PubMed] [Google Scholar]
- 8.Skidmore SJ, Yarbough PO, Gabor KA & Reyes GR Hepatitis E virus: the cause of a waterbourne hepatitis outbreak. J Med Virol 37, 58–60 (1992). [DOI] [PubMed] [Google Scholar]
- 9.Arankalle VA et al. Seroepidemiology of water-borne hepatitis in India and evidence for a third enterically-transmitted hepatitis agent. Proc Natl Acad Sci U S A 91, 3428–3432 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease, C. Enterically transmitted non-A, non-B hepatitis--East Africa. MMWR Morb Mortal Wkly Rep 36, 241–244 (1987). [PubMed] [Google Scholar]
- 11.Leads from the MMWR. Enterically transmitted non-A, non-B hepatitis--East Africa. JAMA 257, 2704–2705 (1987). [PubMed] [Google Scholar]
- 12.Centers for Disease C. Enterically transmitted non-A, non-B hepatitis--Mexico. MMWR Morb Mortal Wkly Rep 36, 597–602 (1987). [PubMed] [Google Scholar]
- 13.Leads from the MMWR. Enterically transmitted non-A, non-B hepatitis--Mexico. JAMA 258, 2036–2037, 2041 (1987). [PubMed] [Google Scholar]
- 14.Iqbal M. et al. An outbreak of enterically transmitted non-A, non-B hepatitis in Pakistan. Am J Trop Med Hyg 40, 438–443 (1989). [DOI] [PubMed] [Google Scholar]
- 15.Fortier D, Treadwell TL & Koff RS Enterically transmitted non-A, non-B hepatitis: importation from Mexico to Massachusetts. N Engl J Med 320, 1281–1282, doi: 10.1056/NEJM198905113201915 (1989). [DOI] [PubMed] [Google Scholar]
- 16.Hlady WG et al. Enterically transmitted non-A, non-B hepatitis associated with an outbreak in Dhaka: epidemiology and public health implications. Trop Doct 20, 15–17, doi: 10.1177/004947559002000105 (1990). [DOI] [PubMed] [Google Scholar]
- 17.Zhuang H, Cao XY, Liu CB & Wang GM Epidemiology of hepatitis E in China. Gastroenterol Jpn 26 Suppl 3, 135–138 (1991). [DOI] [PubMed] [Google Scholar]
- 18.Tsega E. et al. Outbreak of acute hepatitis E virus infection among military personnel in northern Ethiopia. J Med Virol 34, 232–236 (1991). [DOI] [PubMed] [Google Scholar]
- 19.McCarthy MC et al. Acute hepatitis E infection during the 1988 floods in Khartoum, Sudan. Trans R Soc Trop Med Hyg 88, 177 (1994). [DOI] [PubMed] [Google Scholar]
- 20.Guthmann JP et al. A large outbreak of hepatitis E among a displaced population in Darfur, Sudan, 2004: the role of water treatment methods. Clin Infect Dis 42, 1685–1691, doi: 10.1086/504321 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Corwin AL et al. A waterborne outbreak of hepatitis E virus transmission in southwestern Vietnam. Am J Trop Med Hyg 54, 559–562 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Peron JM et al. Hepatitis E is an autochthonous disease in industrialized countries. Analysis of 23 patients in South-West France over a 13-month period and comparison with hepatitis A. Gastroenterol Clin Biol 30, 757–762 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Preiss JC et al. Autochthonous hepatitis E virus infection in Germany with sequence similarities to other European isolates. Infection 34, 173–175, doi: 10.1007/s15010-006-4132-x (2006). [DOI] [PubMed] [Google Scholar]
- 24.Rein DB, Stevens GA, Theaker J, Wittenborn JS & Wiersma ST The global burden of hepatitis E virus genotypes 1 and 2 in 2005. Hepatology 55, 988–997, doi: 10.1002/hep.25505 (2012). [DOI] [PubMed] [Google Scholar]
- 25.WHO. Hepatitis E, <http://www.who.int/mediacentre/factsheets/fs280/en/> (2015). [Google Scholar]
- 26.Ahmad I, Holla RP & Jameel S. Molecular virology of hepatitis E virus. Virus research 161, 47–58, doi: 10.1016/j.virusres.2011.02.011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Cuyck-Gandre H. et al. Short report: polymerase chain reaction detection of hepatitis E virus in north African fecal samples. Am J Trop Med Hyg 54, 134–135 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Escriba JM et al. Hepatitis E, Central African Republic. Emerg Infect Dis 14, 681–683, doi: 10.3201/eid1404.070833 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goumba AI, Konamna X. & Komas NP Clinical and epidemiological aspects of a hepatitis E outbreak in Bangui, Central African Republic. BMC Infect Dis 11, 93, doi: 10.1186/1471-2334-11-93 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hepatitis E, Chad. Wkly Epidemiol Rec 79, 313 (2004). [PubMed] [Google Scholar]
- 31.Coursaget P. et al. Outbreak of enterically-transmitted hepatitis due to hepatitis A and hepatitis E viruses. J Hepatol 28, 745–750 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Ahmed JA et al. Hepatitis E outbreak, Dadaab refugee camp, Kenya, 2012. Emerg Infect Dis 19, 1010–1012, doi: 10.3201/eid1906.130275 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benjelloun S. et al. Seroepidemiological study of an acute hepatitis E outbreak in Morocco. Res Virol 148, 279–287 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Isaacson M, Frean J, He J, Seriwatana J. & Innis BL An outbreak of hepatitis E in Northern Namibia, 1983. Am J Trop Med Hyg 62, 619–625 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Bile K. et al. Contrasting roles of rivers and wells as sources of drinking water on attack and fatality rates in a hepatitis E epidemic in Somalia. Am J Trop Med Hyg 51, 466–474 (1994). [PubMed] [Google Scholar]
- 36.Mushahwar IK, Dawson GJ, Bile KM & Magnius LO Serological studies of an enterically transmitted non-A, non-B hepatitis in Somalia. J Med Virol 40, 218–221 (1993). [DOI] [PubMed] [Google Scholar]
- 37.Centers for Disease, C. & Prevention. Investigation of hepatitis E outbreak among refugees - Upper Nile, South Sudan, 2012–2013. MMWR Morb Mortal Wkly Rep 62, 581–586 (2013). [PMC free article] [PubMed] [Google Scholar]
- 38.Rayis DA, Jumaa AM, Gasim GI, Karsany MS & Adam I. An outbreak of hepatitis E and high maternal mortality at Port Sudan, Eastern Sudan. Pathog Glob Health 107, 66–68, doi: 10.1179/2047773213Y.0000000076 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teshale EH et al. Hepatitis E epidemic, Uganda. Emerg Infect Dis 16, 126–129, doi: 10.3201/eid1601.090764 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clemente-Casares P, Ramos-Romero C, Ramirez-Gonzalez E. & Mas A. Hepatitis E Virus in Industrialized Countries: The Silent Threat. Biomed Res Int 2016, 9838041, doi: 10.1155/2016/9838041 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colson P. et al. Pig liver sausage as a source of hepatitis E virus transmission to humans. The Journal of infectious diseases 202, 825–834, doi: 10.1086/655898 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Lewis HC, Wichmann O. & Duizer E. Transmission routes and risk factors for autochthonous hepatitis E virus infection in Europe: a systematic review. Epidemiology and infection 138, 145–166, doi: 10.1017/s0950268809990847 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Li TC et al. Hepatitis E virus transmission from wild boar meat. Emerging infectious diseases 11, 1958–1960, doi: 10.3201/eid1112.051041 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyamura T. Hepatitis E virus infection in developed countries. Virus research 161, 40–46, doi: 10.1016/j.virusres.2011.03.006 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Ditah I. et al. Current epidemiology of hepatitis E virus infection in the United States: low seroprevalence in the National Health and Nutrition Evaluation Survey. Hepatology 60, 815–822, doi: 10.1002/hep.27219 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Murrison LB & Sherman KE The Enigma of Hepatitis E Virus. Gastroenterol Hepatol (N Y) 13, 484–491 (2017). [PMC free article] [PubMed] [Google Scholar]
- 47.Avellon A, Morago L, Garcia-Galera del Carmen M, Munoz M. & Echevarria JM Comparative sensitivity of commercial tests for hepatitis E genotype 3 virus antibody detection. J Med Virol 87, 1934–1939, doi: 10.1002/jmv.24251 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Vollmer T, Diekmann J, Eberhardt M, Knabbe C. & Dreier J. Monitoring of Anti-Hepatitis E Virus Antibody Seroconversion in Asymptomatically Infected Blood Donors: Systematic Comparison of Nine Commercial Anti-HEV IgM and IgG Assays. Viruses 8, doi: 10.3390/v8080232 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harritshoj LH et al. Low transfusion transmission of hepatitis E among 25,637 single-donation, nucleic acid-tested blood donors. Transfusion 56, 2225–2232, doi: 10.1111/trf.13700 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Gallian P. et al. Hepatitis E virus infections in blood donors, France. Emerg Infect Dis 20, 1914–1917, doi: 10.3201/eid2011.140516 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vollmer T. et al. Novel approach for detection of hepatitis E virus infection in German blood donors. J Clin Microbiol 50, 2708–2713, doi: 10.1128/JCM.01119-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hogema BM et al. Incidence and duration of hepatitis E virus infection in Dutch blood donors. Transfusion 56, 722–728, doi: 10.1111/trf.13402 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Sauleda S. et al. Seroprevalence of hepatitis E virus (HEV) and detection of HEV RNA with a transcription-mediated amplification assay in blood donors from Catalonia (Spain). Transfusion 55, 972–979, doi: 10.1111/trf.12929 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Gallian P. et al. Comparison of hepatitis E virus nucleic acid test screening platforms and RNA prevalence in French blood donors. Transfusion 57, 223–224, doi: 10.1111/trf.13889 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Domanovic D. et al. Hepatitis E and blood donation safety in selected European countries: a shift to screening? Euro Surveill 22, doi: 10.2807/1560-7917.ES.2017.22.16.30514 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roth NJ et al. Low hepatitis E virus RNA prevalence in a large-scale survey of United States source plasma donors. Transfusion, doi: 10.1111/trf.14285 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Meng XJ Expanding Host Range and Cross-Species Infection of Hepatitis E Virus. PLoS Pathog 12, e1005695, doi: 10.1371/journal.ppat.1005695 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoofnagle JH, Nelson KE & Purcell RH Hepatitis E. N Engl J Med 367, 1237–1244, doi: 10.1056/NEJMra1204512 (2012). [DOI] [PubMed] [Google Scholar]
- 59.Meng XJ Zoonotic and foodborne transmission of hepatitis E virus. Semin Liver Dis 33, 41–49, doi: 10.1055/s-0033-1338113 (2013). [DOI] [PubMed] [Google Scholar]
- 60. Kamar N. et al. Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N Engl J Med 358, 811–817, doi: 10.1056/NEJMoa0706992 (2008). It was shown that HEV could become chronic in organ transplant recipients; previously the virus was thought to cause only acute disease.
- 61.Van der Poel WH Food and environmental routes of Hepatitis E virus transmission. Curr Opin Virol 4, 91–96, doi: 10.1016/j.coviro.2014.01.006 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Widen F. et al. Molecular epidemiology of hepatitis E virus in humans, pigs and wild boars in Sweden. Epidemiol Infect 139, 361–371, doi: 10.1017/S0950268810001342 (2011). [DOI] [PubMed] [Google Scholar]
- 63.Jimenez de Oya N. et al. Widespread distribution of hepatitis E virus in Spanish pig herds. BMC Res Notes 4, 412, doi: 10.1186/1756-0500-4-412 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rose N. et al. High prevalence of Hepatitis E virus in French domestic pigs. Comp Immunol Microbiol Infect Dis 34, 419–427, doi: 10.1016/j.cimid.2011.07.003 (2011). [DOI] [PubMed] [Google Scholar]
- 65.Halbur PG et al. Comparative pathogenesis of infection of pigs with hepatitis E viruses recovered from a pig and a human. J Clin Microbiol 39, 918–923, doi: 10.1128/JCM.39.3.918923.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mykytczuk O, Harlow J, Bidawid S, Corneau N. & Nasheri N. Prevalence and Molecular Characterization of the Hepatitis E Virus in Retail Pork Products Marketed in Canada. Food Environ Virol 9, 208–218, doi: 10.1007/s12560-017-9281-9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Szabo K. et al. Detection of hepatitis E virus RNA in raw sausages and liver sausages from retail in Germany using an optimized method. Int J Food Microbiol 215, 149–156, doi: 10.1016/j.ijfoodmicro.2015.09.013 (2015). [DOI] [PubMed] [Google Scholar]
- 68.Pavio N, Merbah T. & Thebault A. Frequent hepatitis E virus contamination in food containing raw pork liver, France. Emerg Infect Dis 20, 1925–1927, doi: 10.3201/eid2011.140891 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barnaud E, Rogee S, Garry P, Rose N. & Pavio N. Thermal inactivation of infectious hepatitis E virus in experimentally contaminated food. Appl Environ Microbiol 78, 5153–5159, doi: 10.1128/AEM.00436-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Woo PC et al. New Hepatitis E Virus Genotype in Bactrian Camels, Xinjiang, China, 2013. Emerg Infect Dis 22, 2219–2221, doi: 10.3201/eid2212.160979 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Woo PC et al. New hepatitis E virus genotype in camels, the Middle East. Emerg Infect Dis 20, 1044–1048, doi: 10.3201/eid2006.140140 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rasche A. et al. Hepatitis E Virus Infection in Dromedaries, North and East Africa, United Arab Emirates, and Pakistan, 1983–2015. Emerg Infect Dis 22, 1249–1252, doi: 10.3201/eid2207.160168 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kumar S, Subhadra S, Singh B. & Panda BK Hepatitis E virus: the current scenario. International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases 17, e228–233, doi: 10.1016/j.ijid.2012.11.026 (2013). [DOI] [PubMed] [Google Scholar]
- 74.Navaneethan U, Al Mohajer M. & Shata MT Hepatitis E and pregnancy: understanding the pathogenesis. Liver Int 28, 1190–1199, doi: 10.1111/j.1478-3231.2008.01840.x (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perez-Gracia MT, Suay-Garcia B. & Mateos-Lindemann ML Hepatitis E and pregnancy: current state. Rev Med Virol, doi: 10.1002/rmv.1929 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Sharapov MB et al. Acute viral hepatitis morbidity and mortality associated with hepatitis E virus infection: Uzbekistan surveillance data. BMC Infect Dis 9, 35, doi: 10.1186/1471-2334-9-35 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pischke S. & Wedemeyer H. Hepatitis E virus infection: multiple faces of an underestimated problem. Journal of hepatology 58, 1045–1046, doi: 10.1016/j.jhep.2012.12.013 (2013). [DOI] [PubMed] [Google Scholar]
- 78.Colson P, Dhiver C, Poizot-Martin I, Tamalet C. & Gerolami R. Acute and chronic hepatitis E in patients infected with human immunodeficiency virus. Journal of viral hepatitis 18, 227–228, doi: 10.1111/j.1365-2893.2010.01311.x (2011). [DOI] [PubMed] [Google Scholar]
- 79.Dalton HR, Bendall RP, Keane FE, Tedder RS & Ijaz S. Persistent carriage of hepatitis E virus in patients with HIV infection. N Engl J Med 361, 1025–1027, doi: 10.1056/NEJMc0903778 (2009). [DOI] [PubMed] [Google Scholar]
- 80.Khuroo MS & Khuroo MS Hepatitis E: an emerging global disease - from discovery towards control and cure. J Viral Hepat 23, 68–79, doi: 10.1111/jvh.12445 (2016). [DOI] [PubMed] [Google Scholar]
- 81.Fujiwara S. et al. Chronic hepatitis E: a review of the literature. J Viral Hepat 21, 78–89, doi: 10.1111/jvh.12156 (2014). [DOI] [PubMed] [Google Scholar]
- 82.Kamar N. et al. Factors associated with chronic hepatitis in patients with hepatitis E virus infection who have received solid organ transplants. Gastroenterology 140, 1481–1489, doi: 10.1053/j.gastro.2011.02.050 (2011). [DOI] [PubMed] [Google Scholar]
- 83.Abravanel F. et al. Hepatitis E virus reinfections in solid-organ-transplant recipients can evolve into chronic infections. J Infect Dis 209, 1900–1906, doi: 10.1093/infdis/jiu032 (2014). [DOI] [PubMed] [Google Scholar]
- 84.Borentain P. et al. Hepatocellular carcinoma complicating hepatitis E virus-related cirrhosis. Hepatology, doi: 10.1002/hep.29508 (2017). [DOI] [PubMed] [Google Scholar]
- 85.Kamar N. et al. Hepatitis E virus-induced neurological symptoms in a kidney-transplant patient with chronic hepatitis. Am J Transplant 10, 1321–1324, doi: 10.1111/j.1600-6143.2010.03068.x (2010). [DOI] [PubMed] [Google Scholar]
- 86.van den Berg B. et al. Guillain-Barre syndrome associated with preceding hepatitis E virus infection. Neurology 82, 491–497, doi: 10.1212/WNL.0000000000000111 (2014). [DOI] [PubMed] [Google Scholar]
- 87.Bazerbachi F, Haffar S, Garg SK & Lake JR Extra-hepatic manifestations associated with hepatitis E virus infection: a comprehensive review of the literature. Gastroenterol Rep (Oxf) 4, 1–15, doi: 10.1093/gastro/gov042 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pischke S. et al. Hepatitis E virus: Infection beyond the liver? J Hepatol 66, 1082–1095, doi: 10.1016/j.jhep.2016.11.016 (2017). [DOI] [PubMed] [Google Scholar]
- 89.McLean BN, Gulliver J. & Dalton HR Hepatitis E virus and neurological disorders. Pract Neurol, doi: 10.1136/practneurol-2016-001588 (2017). [DOI] [PubMed] [Google Scholar]
- 90.Mallet V. et al. Hepatitis E Virus-Induced Primary Cutaneous CD30(+) T cell Lymphoproliferative Disorder. J Hepatol, doi: 10.1016/j.jhep.2017.08.011 (2017). [DOI] [PubMed] [Google Scholar]
- 91.Comont T. et al. [Acute hepatitis E infection associated with Guillain-Barre syndrome in an immunocompetent patient]. Rev Med Interne 35, 333–336, doi: 10.1016/j.revmed.2013.05.005 (2014). [DOI] [PubMed] [Google Scholar]
- 92.Zhou X. et al. Hepatitis E Virus Infects Neurons and Brains. J Infect Dis 215, 1197–1206, doi: 10.1093/infdis/jix079 (2017). [DOI] [PubMed] [Google Scholar]
- 93.Choi C. & Chae C. Localization of swine hepatitis E virus in liver and extrahepatic tissues from naturally infected pigs by in situ hybridization. J Hepatol 38, 827–832 (2003). [DOI] [PubMed] [Google Scholar]
- 94.Kamar N. et al. Influence of immunosuppressive therapy on the natural history of genotype 3 hepatitis-E virus infection after organ transplantation. Transplantation 89, 353–360, doi: 10.1097/TP.0b013e3181c4096c (2010). [DOI] [PubMed] [Google Scholar]
- 95. Kamar N. et al. Ribavirin for chronic hepatitis E virus infection in transplant recipients. N Engl J Med 370, 1111–1120, doi: 10.1056/NEJMoa1215246 (2014). Ribavirin monotherapy led to HEV clearance in 95% of patients in a cohort of chronically infected solid-organ transplant recipients.
- 96.Khuroo MS & Kamili S. Aetiology, clinical course and outcome of sporadic acute viral hepatitis in pregnancy. J Viral Hepat 10, 61–69 (2003). [DOI] [PubMed] [Google Scholar]
- 97. Debing Y. et al. A mutation in the hepatitis E virus RNA polymerase promotes its replication and associates with ribavirin treatment failure in organ transplant recipients. Gastroenterology 147, 1008–1011 e1007; quiz e1015–1006, doi: 10.1053/j.gastro.2014.08.040 (2014). A mutation in the polymerase of HEV was discovered in two patients who were nonresponsive to ribavirin therapy. In vitro, this mutation increased replication efficiency of the virus.
- 98.Debing Y. et al. Hepatitis E virus mutations associated with ribavirin treatment failure result in altered viral fitness and ribavirin sensitivity. J Hepatol 65, 499–508, doi: 10.1016/j.jhep.2016.05.002 (2016). [DOI] [PubMed] [Google Scholar]
- 99.Mazzola A. et al. Chronic hepatitis E viral infection after liver transplantation: a regression of fibrosis following antiviral therapy. Transplantation, doi: 10.1097/TP.0000000000001766 (2017). [DOI] [PubMed] [Google Scholar]
- 100.Alric L, Bonnet D, Laurent G, Kamar N. & Izopet J. Chronic hepatitis E virus infection: successful virologic response to pegylated interferon-alpha therapy. Ann Intern Med 153, 135136, doi: 10.7326/0003-4819-153-2-201007200-00256 (2010). [DOI] [PubMed] [Google Scholar]
- 101.Kamar N. et al. Three-month pegylated interferon-alpha-2a therapy for chronic hepatitis E virus infection in a haemodialysis patient. Nephrol Dial Transplant 25, 2792–2795, doi: 10.1093/ndt/gfq282 (2010). [DOI] [PubMed] [Google Scholar]
- 102.Dao Thi VL et al. Sofosbuvir Inhibits Hepatitis E Virus Replication In Vitro and Results in an Additive Effect When Combined With Ribavirin. Gastroenterology 150, 82–85 e84, doi: 10.1053/j.gastro.2015.09.011 (2016). [DOI] [PubMed] [Google Scholar]
- 103.van der Valk M, Zaaijer HL, Kater AP & Schinkel J. Sofosbuvir shows antiviral activity in a patient with chronic hepatitis E virus infection. J Hepatol 66, 242–243, doi: 10.1016/j.jhep.2016.09.014 (2017). [DOI] [PubMed] [Google Scholar]
- 104.Todt D. et al. In vivo evidence for ribavirin-induced mutagenesis of the hepatitis E virus genome. Gut 65, 1733–1743, doi: 10.1136/gutjnl-2015-311000 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lhomme S. et al. Mutation in the Hepatitis E Virus Polymerase and Outcome of Ribavirin Therapy. Antimicrob Agents Chemother 60, 1608–1614, doi: 10.1128/AAC.02496-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhao Q. et al. Antigenic determinants of hepatitis E virus and vaccine-induced immunogenicity and efficacy. J Gastroenterol 48, 159–168, doi: 10.1007/s00535-012-0701-1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lee GY et al. Hepatitis E virus infection: Epidemiology and treatment implications. World journal of virology 4, 343–355, doi: 10.5501/wjv.v4.i4.343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wu T. et al. Difference of T cell and B cell activation in two homologous proteins with similar antigenicity but great distinct immunogenicity. Mol Immunol 44, 3261–3266, doi: 10.1016/j.molimm.2007.01.002 (2007). [DOI] [PubMed] [Google Scholar]
- 109.Tsarev SA et al. Recombinant vaccine against hepatitis E: dose response and protection against heterologous challenge. Vaccine 15, 1834–1838 (1997). [DOI] [PubMed] [Google Scholar]
- 110.Shrestha MP et al. Safety and efficacy of a recombinant hepatitis E vaccine. N Engl J Med 356, 895–903, doi: 10.1056/NEJMoa061847 (2007). [DOI] [PubMed] [Google Scholar]
- 111. Zhu FC et al. Efficacy and safety of a recombinant hepatitis E vaccine in healthy adults: a large-scale, randomised, double-blind placebo-controlled, phase 3 trial. Lancet 376, 895–902, doi: 10.1016/S0140-6736(10)61030-6 (2010). The HEV239 vaccine was found to be 100% effective (95% CI 72.1–100) after three doses in a phase 3 clinical trial in China.
- 112.Joshi SS & Arankalle VA Enhanced humoral response in pregnant mice immunized with liposome encapsulated recombinant neutralizing epitope protein of Hepatitis- E virus. Virol J 12, 70, doi: 10.1186/s12985-015-0302-8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vaccines prequalification priority list 2015–16, <http://www.who.int/immunization_standards/vaccine_quality/priority_pq_vaccines_2015_16/en/> (2016). [Google Scholar]
- 114. Guu TS et al. Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding. Proc Natl Acad Sci U S A 106, 12992–12997, doi: 10.1073/pnas.0904848106 (2009). The crystal structure of the HEV virus-like particle, composed of self-assembling capsid proteins, was determined to a 3.5Å resolution.
- 115.Feng Z. et al. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496, 367–371, doi: 10.1038/nature12029 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Takahashi M. et al. Monoclonal antibodies raised against the ORF3 protein of hepatitis E virus (HEV) can capture HEV particles in culture supernatant and serum but not those in feces. Arch Virol 153, 1703–1713, doi: 10.1007/s00705-008-0179-6 (2008). It was first shown that HEV particles in culture supernatant and feces have distinct properties.
- 117. Yin X, Ambardekar C, Lu Y. & Feng Z. Distinct Entry Mechanisms for Nonenveloped and Quasi-Enveloped Hepatitis E Viruses. J Virol 90, 4232–4242, doi: 10.1128/JVI.02804-15 (2016). HEV was formally stated to be a quasi-enveloped virus, with the enveloped and nonenveloped forms having different entry mechanisms.
- 118.Chapuy-Regaud S. et al. Characterization of the lipid envelope of exosome encapsulated HEV particles protected from the immune response. Biochimie, doi: 10.1016/j.biochi.2017.05.003 (2017). [DOI] [PubMed] [Google Scholar]
- 119.Nagashima S. et al. The characterization of the quasi-enveloped hepatitis E virus particles released by the cellular exosomal pathway. J Virol, doi: 10.1128/JVI.00822-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang M, Purcell RH & Emerson SU Identification of the 5’ terminal sequence of the SAR-55 and MEX-14 strains of hepatitis E virus and confirmation that the genome is capped. J Med Virol 65, 293–295 (2001). [DOI] [PubMed] [Google Scholar]
- 121.Tam AW et al. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology 185, 120–131 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nair VP et al. Endoplasmic Reticulum Stress Induced Synthesis of a Novel Viral Factor Mediates Efficient Replication of Genotype-1 Hepatitis E Virus. PLoS Pathog 12, e1005521, doi: 10.1371/journal.ppat.1005521 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fry KE et al. Hepatitis E virus (HEV): strain variation in the nonstructural gene region encoding consensus motifs for an RNA-dependent RNA polymerase and an ATP/GTP binding site. Virus Genes 6, 173–185 (1992). [DOI] [PubMed] [Google Scholar]
- 124.Rozanov MN, Koonin EV & Gorbalenya AE Conservation of the putative methyltransferase domain: a hallmark of the ‘Sindbis-like’ supergroup of positive-strand RNA viruses. J Gen Virol 73 ( Pt 8), 2129–2134, doi: 10.1099/0022-1317-73-8-2129 (1992). [DOI] [PubMed] [Google Scholar]
- 125.Magden J. et al. Virus-specific mRNA capping enzyme encoded by hepatitis E virus. J Virol 75, 6249–6255, doi: 10.1128/JVI.75.14.6249-6255.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Koonin EV et al. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc Natl Acad Sci U S A 89, 8259–8263 (1992). It was predicted based on sequence homology that HEV contains a domain similar to the papain-like cysteine protease of rubella virus, however to date protease function has not been conclusively proven for HEV.
- 127.Chen JP, Strauss JH, Strauss EG & Frey TK Characterization of the rubella virus nonstructural protease domain and its cleavage site. J Virol 70, 4707–4713 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Golubtsov A, Kaariainen L. & Caldentey J. Characterization of the cysteine protease domain of Semliki Forest virus replicase protein nsP2 by in vitro mutagenesis. FEBS Lett 580, 1502–1508, doi: 10.1016/j.febslet.2006.01.071 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jore J, De Geus B, Jackson RJ, Pouwels PH & Enger-Valk BE Poliovirus protein 3CD is the active protease for processing of the precursor protein P1 in vitro. J Gen Virol 69 ( Pt 7), 1627–1636, doi: 10.1099/0022-1317-69-7-1627 (1988). [DOI] [PubMed] [Google Scholar]
- 130.Jameel S, Zafrullah M, Ozdener MH & Panda SK Expression in animal cells and characterization of the hepatitis E virus structural proteins. J Virol 70, 207–216 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Ding Q. et al. Hepatitis E virus ORF3 is a functional ion channel required for release of infectious particles. Proc Natl Acad Sci U S A 114, 1147–1152, doi: 10.1073/pnas.1614955114 (2017). HEV ORF3, a viral factor critical for release, was shown to exhibit ion channel activity; therefore HEV is now known to contain a viroporin.
- 132. Nagashima S. et al. A PSAP motif in the ORF3 protein of hepatitis E virus is necessary for virion release from infected cells. J Gen Virol 92, 269–278, doi: 10.1099/vir.0.025791-0 (2011). A specific PSAP motif within the HEV ORF3 protein is involved in viral release.
- 133.Martin-Serrano J, Zang T. & Bieniasz PD Role of ESCRT-I in retroviral budding. J Virol 77, 4794–4804 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kalia M, Chandra V, Rahman SA, Sehgal D. & Jameel S. Heparan sulfate proteoglycans are required for cellular binding of the hepatitis E virus ORF2 capsid protein and for viral infection. J Virol 83, 12714–12724, doi: 10.1128/JVI.00717-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kapur N, Thakral D, Durgapal H. & Panda SK Hepatitis E virus enters liver cells through receptor-dependent clathrin-mediated endocytosis. J Viral Hepat 19, 436–448, doi: 10.1111/j.1365-2893.2011.01559.x (2012). [DOI] [PubMed] [Google Scholar]
- 136. Holla P, Ahmad I, Ahmed Z. & Jameel S. Hepatitis E virus enters liver cells through a dynamin-2, clathrin and membrane cholesterol-dependent pathway. Traffic 16, 398–416, doi: 10.1111/tra.12260 (2015). Little was known about host factors involved in HEV entry until this study, which used specific small molecule inhibitors to identify cellular proteins mediating entry of fluorescent HEV-like particles.
- 137. Graff J, Torian U, Nguyen H. & Emerson SU A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E virus. J Virol 80, 5919–5926, doi: 10.1128/JVI.00046-06 (2006). This study provided the first evidence that HEV ORF2 and ORF3 proteins are translated from a subgenomic RNA.
- 138.Agrawal S, Gupta D. & Panda SK The 3’ end of hepatitis E virus (HEV) genome binds specifically to the viral RNA-dependent RNA polymerase (RdRp). Virology 282, 87–101, doi: 10.1006/viro.2000.0819 (2001). [DOI] [PubMed] [Google Scholar]
- 139. Nanda SK, Panda SK, Durgapal H. & Jameel S. Detection of the negative strand of hepatitis E virus RNA in the livers of experimentally infected rhesus monkeys: evidence for viral replication. J Med Virol 42, 237–240 (1994). HEV can replicate in the livers of rhesus monkeys. This was proven through the detection of negative-strand RNA, a technique that was later used to validate other animal models for HEV, and to discover extrahepatic sites of HEV replication.
- 140.Varma SP et al. Hepatitis E virus replication involves alternating negative- and positive-sense RNA synthesis. J Gen Virol 92, 572–581, doi: 10.1099/vir.0.027714-0 (2011). [DOI] [PubMed] [Google Scholar]
- 141.Xia X, Huang R. & Li D. [Studies on the subgenomic RNAs of hepatitis E virus]. Wei Sheng Wu Xue Bao 40, 622–627 (2000). [PubMed] [Google Scholar]
- 142.Yarbough PO et al. Hepatitis E virus: identification of type-common epitopes. J Virol 65, 5790–5797 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mahilkar S, Paingankar MS & Lole KS Hepatitis E virus RNA-dependent RNA polymerase: RNA template specificities, recruitment and synthesis. J Gen Virol 97, 2231–2242, doi: 10.1099/jgv.0.000528 (2016). [DOI] [PubMed] [Google Scholar]
- 144.Zafrullah M, Ozdener MH, Kumar R, Panda SK & Jameel S. Mutational analysis of glycosylation, membrane translocation, and cell surface expression of the hepatitis E virus ORF2 protein. J Virol 73, 4074–4082 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Graff J. et al. Mutations within potential glycosylation sites in the capsid protein of hepatitis E virus prevent the formation of infectious virus particles. J Virol 82, 1185–1194, doi: 10.1128/JVI.01219-07 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Surjit M, Jameel S. & Lal SK The ORF2 protein of hepatitis E virus binds the 5’ region of viral RNA. J Virol 78, 320–328 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Surjit M, Jameel S. & Lal SK Cytoplasmic localization of the ORF2 protein of hepatitis E virus is dependent on its ability to undergo retrotranslocation from the endoplasmic reticulum. J Virol 81, 3339–3345, doi: 10.1128/JVI.02039-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lenggenhager D. et al. Visualization of hepatitis E virus RNA and proteins in the human liver. J Hepatol, doi: 10.1016/j.jhep.2017.04.002 (2017). [DOI] [PubMed] [Google Scholar]
- 149.Suzuki T. et al. The human polyoma JC virus agnoprotein acts as a viroporin. PLoS Pathog 6, e1000801, doi: 10.1371/journal.ppat.1000801 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nieva JL, Madan V. & Carrasco L. Viroporins: structure and biological functions. Nat Rev Microbiol 10, 563–574, doi: 10.1038/nrmicro2820 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zafrullah M, Ozdener MH, Panda SK & Jameel S. The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J Virol 71, 9045–9053 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Korkaya H. et al. The ORF3 protein of hepatitis E virus binds to Src homology 3 domains and activates MAPK. J Biol Chem 276, 42389–42400, doi: 10.1074/jbc.M101546200 (2001). [DOI] [PubMed] [Google Scholar]
- 153.Parvez MK & Al-Dosari MS Evidence of MAPK-JNK1/2 activation by hepatitis E virus ORF3 protein in cultured hepatoma cells. Cytotechnology 67, 545–550, doi: 10.1007/s10616-014-9785-1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kar-Roy A, Korkaya H, Oberoi R, Lal SK & Jameel S. The hepatitis E virus open reading frame 3 protein activates ERK through binding and inhibition of the MAPK phosphatase. J Biol Chem 279, 28345–28357, doi: 10.1074/jbc.M400457200 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tyagi S, Surjit M. & Lal SK The 41-amino-acid C-terminal region of the hepatitis E virus ORF3 protein interacts with bikunin, a kunitz-type serine protease inhibitor. J Virol 79, 1208112087, doi: 10.1128/JVI.79.18.12081-12087.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ratra R, Kar-Roy A. & Lal SK The ORF3 protein of hepatitis E virus interacts with hemopexin by means of its 26 amino acid N-terminal hydrophobic domain II. Biochemistry 47, 1957–1969, doi: 10.1021/bi7016552 (2008). [DOI] [PubMed] [Google Scholar]
- 157.Ratra R, Kar-Roy A. & Lal SK ORF3 protein of hepatitis E virus interacts with the Bbeta chain of fibrinogen resulting in decreased fibrinogen secretion from HuH-7 cells. J Gen Virol 90, 13591370, doi: 10.1099/vir.0.009274-0 (2009). [DOI] [PubMed] [Google Scholar]
- 158.Kannan H, Fan S, Patel D, Bossis I. & Zhang YJ The hepatitis E virus open reading frame 3 product interacts with microtubules and interferes with their dynamics. J Virol 83, 6375–6382, doi: 10.1128/JVI.02571-08 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Moin SM, Chandra V, Arya R. & Jameel S. The hepatitis E virus ORF3 protein stabilizes HIF-1alpha and enhances HIF-1-mediated transcriptional activity through p300/CBP. Cell Microbiol 11, 1409–1421, doi: 10.1111/j.1462-5822.2009.01340.x (2009). [DOI] [PubMed] [Google Scholar]
- 160.Chandra V, Kalia M, Hajela K. & Jameel S. The ORF3 protein of hepatitis E virus delays degradation of activated growth factor receptors by interacting with CIN85 and blocking formation of the Cbl-CIN85 complex. J Virol 84, 3857–3867, doi: 10.1128/JVI.01994-09 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chandra V. et al. The hepatitis E virus ORF3 protein regulates the expression of liver-specific genes by modulating localization of hepatocyte nuclear factor 4. PLoS One 6, e22412, doi: 10.1371/journal.pone.0022412 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wang C. et al. HEV-ORF3 Encoding Phosphoprotein Interacts With Hepsin. Hepat Mon 14, e13902, doi: 10.5812/hepatmon.13902 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chandra V, Kar-Roy A, Kumari S, Mayor S. & Jameel S. The hepatitis E virus ORF3 protein modulates epidermal growth factor receptor trafficking, STAT3 translocation, and the acute-phase response. J Virol 82, 7100–7110, doi: 10.1128/JVI.00403-08 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nagashima S. et al. The membrane on the surface of hepatitis E virus particles is derived from the intracellular membrane and contains trans-Golgi network protein 2. Arch Virol 159, 979–991, doi: 10.1007/s00705-013-1912-3 (2014). [DOI] [PubMed] [Google Scholar]
- 165.Surjit M, Oberoi R, Kumar R. & Lal SK Enhanced alpha1 microglobulin secretion from Hepatitis E virus ORF3-expressing human hepatoma cells is mediated by the tumor susceptibility gene 101. J Biol Chem 281, 8135–8142, doi: 10.1074/jbc.M509568200 (2006). [DOI] [PubMed] [Google Scholar]
- 166.Yamada K. et al. ORF3 protein of hepatitis E virus is essential for virion release from infected cells. J Gen Virol 90, 1880–1891, doi: 10.1099/vir.0.010561-0 (2009). [DOI] [PubMed] [Google Scholar]
- 167.Emerson SU et al. Release of genotype 1 hepatitis E virus from cultured hepatoma and polarized intestinal cells depends on open reading frame 3 protein and requires an intact PXXP motif. J Virol 84, 9059–9069, doi: 10.1128/JVI.00593-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kenney SP, Wentworth JL, Heffron CL & Meng XJ Replacement of the hepatitis E virus ORF3 protein PxxP motif with heterologous late domain motifs affects virus release via interaction with TSG101. Virology 486, 198–208, doi: 10.1016/j.virol.2015.09.012 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nagashima S. et al. Hepatitis E virus egress depends on the exosomal pathway, with secretory exosomes derived from multivesicular bodies. J Gen Virol 95, 2166–2175, doi: 10.1099/vir.0.066910-0 (2014). [DOI] [PubMed] [Google Scholar]
- 170.Qi Y. et al. Hepatitis E Virus Produced from Cell Culture Has a Lipid Envelope. PLoS One 10, e0132503, doi: 10.1371/journal.pone.0132503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Paingankar MS & Arankalle VA Identification and characterization of cellular proteins interacting with Hepatitis E virus untranslated regions. Virus Res 208, 98–109, doi: 10.1016/j.virusres.2015.06.006 (2015). [DOI] [PubMed] [Google Scholar]
- 172.Hurley JH The ESCRT complexes. Crit Rev Biochem Mol Biol 45, 463–487, doi: 10.3109/10409238.2010.502516 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Tam AW et al. In vitro infection and replication of hepatitis E virus in primary cynomolgus macaque hepatocytes. Virology 238, 94–102, doi: 10.1006/viro.1997.8817 (1997). This was the first use of primary hepatocytes to study HEV in vitro.
- 174.Panda SK, Ansari IH, Durgapal H, Agrawal S. & Jameel S. The in vitro-synthesized RNA from a cDNA clone of hepatitis E virus is infectious. J Virol 74, 2430–2437 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Huang R. et al. Cell culture of sporadic hepatitis E virus in China. Clin Diagn Lab Immunol 6, 729–733 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kazachkov Yu A. et al. Hepatitis E virus in cultivated cells. Arch Virol 127, 399–402 (1992). [DOI] [PubMed] [Google Scholar]
- 177.Tanaka T. et al. Development and characterization of a genotype 4 hepatitis E virus cell culture system using a HE-JF5/15F strain recovered from a fulminant hepatitis patient. J Clin Microbiol 47, 1906–1910, doi:1 10.1128/JCM.00629-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yamada K. et al. Construction of an infectious cDNA clone of hepatitis E virus strain JE03–1760F that can propagate efficiently in cultured cells. J Gen Virol 90, 457–462, doi: 10.1099/vir.0.0075590 (2009). [DOI] [PubMed] [Google Scholar]
- 179. Shukla P. et al. Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proc Natl Acad Sci U S A 108, 2438–2443, doi: 10.1073/pnas.1018878108 (2011). A cell-culture adapted HEV strain, Kernow-C1/p6, was discovered and found to contain a human ribosomal insert that expanded its host range and led to more efficient replication. This HEV strain has been widely used for subsequent in vitro studies.
- 180.Shukla P. et al. Adaptation of a genotype 3 hepatitis E virus to efficient growth in cell culture depends on an inserted human gene segment acquired by recombination. J Virol 86, 5697–5707, doi: 10.1128/JVI.00146-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Nguyen HT et al. A naturally occurring human/hepatitis E recombinant virus predominates in serum but not in faeces of a chronic hepatitis E patient and has a growth advantage in cell culture. J Gen Virol 93, 526–530, doi: 10.1099/vir.0.037259-0 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yin X. et al. Hepatitis E virus persists in the presence of a type III interferon response. PLoS Pathog 13, e1006417, doi: 10.1371/journal.ppat.1006417 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Xu L. et al. RIG-I Is A Key Antiviral Interferon-Stimulated Gene Against Hepatitis E Virus Dispensable Of Interferon Production. Hepatology, doi: 10.1002/hep.29105 (2017). [DOI] [PubMed] [Google Scholar]
- 184.Rogee S. et al. New models of hepatitis E virus replication in human and porcine hepatocyte cell lines. J Gen Virol 94, 549–558, doi: 10.1099/vir.0.049858-0 (2013). [DOI] [PubMed] [Google Scholar]
- 185.Elkayam T, Amitay-Shaprut S, Dvir-Ginzberg M, Harel T. & Cohen S. Enhancing the drug metabolism activities of C3A--a human hepatocyte cell line--by tissue engineering within alginate scaffolds. Tissue Eng 12, 1357–1368, doi: 10.1089/ten.2006.12.1357 (2006). [DOI] [PubMed] [Google Scholar]
- 186.Dembele L. et al. Persistence and activation of malaria hypnozoites in long-term primary hepatocyte cultures. Nature medicine 20, 307–312, doi: 10.1038/nm.3461 (2014). [DOI] [PubMed] [Google Scholar]
- 187.Mitaka T. The current status of primary hepatocyte culture. Int J Exp Pathol 79, 393–409 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Thomas E. & Liang TJ Experimental models of hepatitis B and C - new insights and progress. Nature reviews. Gastroenterology & hepatology 13, 362–374, doi: 10.1038/nrgastro.2016.37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Verrier ER, Colpitts CC, Schuster C, Zeisel MB & Baumert TF Cell Culture Models for the Investigation of Hepatitis B and D Virus Infection. Viruses 8, doi: 10.3390/v8090261 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Dunn JC, Tompkins RG & Yarmush ML Long-term in vitro function of adult hepatocytes in a collagen sandwich configuration. Biotechnol Prog 7, 237–245, doi: 10.1021/bp00009a007 (1991). [DOI] [PubMed] [Google Scholar]
- 191.Dunn JC, Yarmush ML, Koebe HG & Tompkins RG Hepatocyte function and extracellular matrix geometry: long-term culture in a sandwich configuration. FASEB J 3, 174–177 (1989). [DOI] [PubMed] [Google Scholar]
- 192.Guillouzo A, Morel F, Ratanasavanh D, Chesne C. & Guguen-Guillouzo C. Long-term culture of functional hepatocytes. Toxicol In Vitro 4, 415–427 (1990). [DOI] [PubMed] [Google Scholar]
- 193.Chong TW et al. Primary human hepatocytes in spheroid formation to study hepatitis C infection. J Surg Res 130, 52–57, doi: 10.1016/j.jss.2005.04.043 (2006). [DOI] [PubMed] [Google Scholar]
- 194.Kidambi S. et al. Oxygen-mediated enhancement of primary hepatocyte metabolism, functional polarization, gene expression, and drug clearance. Proc Natl Acad Sci U S A 106, 15714–15719, doi:0906820106 [pii] 10.1073/pnas.0906820106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Khetani SR & Bhatia SN Microscale culture of human liver cells for drug development. Nat Biotechnol 26, 120–126, doi:nbt1361 [pii] 10.1038/nbt1361 (2008). [DOI] [PubMed] [Google Scholar]
- 196.Bhatia SN, Balis UJ, Yarmush ML & Toner M. Effect of cell-cell interactions in preservation of cellular phenotype: cocultivation of hepatocytes and nonparenchymal cells. FASEB J 13, 1883–1900 (1999). [DOI] [PubMed] [Google Scholar]
- 197.Shlomai A. et al. Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell-derived hepatocellular systems. Proc Natl Acad Sci U S A 111, 1219312198, doi: 10.1073/pnas.1412631111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.March S. et al. A Microscale Human Liver Platform that Supports the Hepatic Stages of Plasmodium falciparum and vivax. Cell Host Microbe 14, 104–115, doi: 10.1016/j.chom.2013.06.005 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ploss A. et al. Persistent hepatitis C virus infection in microscale primary human hepatocyte cultures. Proceedings of the National Academy of Sciences of the United States of America 107, 3141–3145, doi: 10.1073/pnas.0915130107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Winer BY et al. Long-term hepatitis B infection in a scalable hepatic co-culture system. Nat Commun 8, 125, doi: 10.1038/s41467-017-00200-8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Helsen N. et al. Stem cell-derived hepatocytes: A novel model for hepatitis E virus replication. J Hepatol 64, 565–573, doi: 10.1016/j.jhep.2015.11.013 (2016). Human pluripotent stem cell-derived hepatocyte-like cells were established as an in vitro model to study HEV.
- 202.Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell 10, 678684, doi: 10.1016/j.stem.2012.05.005 (2012). [DOI] [PubMed] [Google Scholar]
- 203.Ramanan V, Scull MA, Sheahan TP, Rice CM & Bhatia SN New Methods in Tissue Engineering: Improved Models for Viral Infection. Annu Rev Virol 1, 475–499, doi: 10.1146/annurev-virology-031413-085437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Irion S, Nostro MC, Kattman SJ & Keller GM Directed differentiation of pluripotent stem cells: from developmental biology to therapeutic applications. Cold Spring Harb Symp Quant Biol 73, 101–110, doi: 10.1101/sqb.2008.73.065 (2008). [DOI] [PubMed] [Google Scholar]
- 205.Davidson MD, Kukla DA & Khetani SR Microengineered cultures containing human hepatic stellate cells and hepatocytes for drug development. Integr Biol (Camb) 9, 662–677, doi: 10.1039/c7ib00027h (2017). [DOI] [PubMed] [Google Scholar]
- 206.Ma X. et al. Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc Natl Acad Sci U S A 113, 2206–2211, doi: 10.1073/pnas.1524510113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chen AA et al. Humanized mice with ectopic artificial liver tissues. Proc Natl Acad Sci U S A 108, 11842–11847, doi: 10.1073/pnas.1101791108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.March S. et al. Micropatterned coculture of primary human hepatocytes and supportive cells for the study of hepatotropic pathogens. Nat Protoc 10, 2027–2053, doi: 10.1038/nprot.2015.128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bose PD et al. Evidence of extrahepatic replication of hepatitis E virus in human placenta. J Gen Virol 95, 1266–1271, doi: 10.1099/vir.0.063602-0 (2014). [DOI] [PubMed] [Google Scholar]
- 210.de Deus N. et al. Detection of hepatitis E virus in liver, mesenteric lymph node, serum, bile and faeces of naturally infected pigs affected by different pathological conditions. Vet Microbiol 119, 105–114, doi: 10.1016/j.vetmic.2006.08.027 (2007). [DOI] [PubMed] [Google Scholar]
- 211.Williams TP et al. Evidence of extrahepatic sites of replication of the hepatitis E virus in a swine model. J Clin Microbiol 39, 3040–3046 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Zhou X. et al. Hepatitis E virus infects neurons and brains. J Infect Dis, doi: 10.1093/infdis/jix079 (2017). [DOI] [PubMed] [Google Scholar]
- 213.Huang R. et al. Hepatitis E virus (87A strain) propagated in A549 cells. J Med Virol 47, 299–302 (1995). [DOI] [PubMed] [Google Scholar]
- 214.Takahashi H. et al. A549 and PLC/PRF/5 cells can support the efficient propagation of swine and wild boar hepatitis E virus (HEV) strains: demonstration of HEV infectivity of porcine liver sold as food. Arch Virol 157, 235–246, doi: 10.1007/s00705-011-1153-2 (2012). [DOI] [PubMed] [Google Scholar]
- 215.Jirintai S. et al. Molecular analysis of hepatitis E virus from farm rabbits in Inner Mongolia, China and its successful propagation in A549 and PLC/PRF/5 cells. Virus Res 170, 126–137, doi: 10.1016/j.virusres.2012.09.015 (2012). [DOI] [PubMed] [Google Scholar]
- 216.Montalvo Villalba MC et al. Hepatitis E virus in bottlenose dolphins Tursiops truncatus. Dis Aquat Organ 123, 13–18, doi: 10.3354/dao03085 (2017). [DOI] [PubMed] [Google Scholar]
- 217.Panda SK, Datta R, Kaur J, Zuckerman AJ & Nayak NC Enterically transmitted non-A, non-B hepatitis: recovery of virus-like particles from an epidemic in south Delhi and transmission studies in rhesus monkeys. Hepatology 10, 466–472 (1989). [DOI] [PubMed] [Google Scholar]
- 218.Uchida T. et al. Animal model, virology and gene cloning of hepatitis E. Gastroenterol Jpn 26 Suppl 3, 148–151 (1991). [DOI] [PubMed] [Google Scholar]
- 219.Arankalle VA, Chadha MS & Chobe LP Long-term serological follow up and crosschallenge studies in rhesus monkeys experimentally infected with hepatitis E virus. J Hepatol 30, 199–204 (1999). [DOI] [PubMed] [Google Scholar]
- 220. Yu C. et al. Pathogenesis of hepatitis E virus and hepatitis C virus in chimpanzees: similarities and differences. J Virol 84, 11264–11278, doi: 10.1128/JVI.01205-10 (2010). The authors compared host responses to HCV and HEV in chimpanzees, which are the only non-human host for both viruses.
- 221.Shata MT et al. Characterization of hepatitis E-specific cell-mediated immune response using IFN-gamma ELISPOT assay. J Immunol Methods 328, 152–161, doi: 10.1016/j.jim.2007.08.014 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Li X, Kamili S. & Krawczynski K. Quantitative detection of hepatitis E virus RNA and dynamics of viral replication in experimental infection. J Viral Hepat 13, 835–839, doi: 10.1111/j.13652893.2006.00754.x (2006). [DOI] [PubMed] [Google Scholar]
- 223. Emerson SU et al. Recombinant hepatitis E virus genomes infectious for primates: importance of capping and discovery of a cis-reactive element. Proc Natl Acad Sci U S A 98, 15270–15275, doi: 10.1073/pnas.251555098 (2001). The authors infected rhesus monkeys and chimpanzees with capped versus uncapped HEV transcripts, and found improved infection efficiency and more severe clinical phenotype with the capped transcript.
- 224. Meng XJ et al. Genetic and experimental evidence for cross-species infection by swine hepatitis E virus. J Virol 72, 9714–9721 (1998). HEV from humans was capable of infecting swine, and sequence analysis revealed that human and swine HEV strains in the United States were similar. This data strongly suggested the possibility of zoonotic transmission of HEV from swine to humans, which is now known to be the primary cause of HEV in developed nations.
- 225.Krawczynski K, Meng XJ & Rybczynska J. Pathogenetic elements of hepatitis E and animal models of HEV infection. Virus Res 161, 78–83, doi: 10.1016/j.virusres.2011.03.007 (2011). [DOI] [PubMed] [Google Scholar]
- 226.Tsarev SA et al. Variation in course of hepatitis E in experimentally infected cynomolgus monkeys. J Infect Dis 167, 1302–1306 (1993). [DOI] [PubMed] [Google Scholar]
- 227.Tsarev SA et al. Infectivity titration of a prototype strain of hepatitis E virus in cynomolgus monkeys. J Med Virol 43, 135–142 (1994). [DOI] [PubMed] [Google Scholar]
- 228.Soe S. et al. Enterically transmitted non-A, non-B hepatitis in cynomolgus monkeys: morphology and probable mechanism of hepatocellular necrosis. Liver 9, 135–145 (1989). [DOI] [PubMed] [Google Scholar]
- 229.Tsarev SA et al. Successful passive and active immunization of cynomolgus monkeys against hepatitis E. Proc Natl Acad Sci U S A 91, 10198–10202 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Arankalle VA, Chadha MS, Chobe LP, Nair R. & Banerjee K. Cross-challenge studies in rhesus monkeys employing different Indian isolates of hepatitis E virus. J Med Virol 46, 358–363 (1995). [DOI] [PubMed] [Google Scholar]
- 231.Zhang M. et al. Recombinant vaccine against hepatitis E: duration of protective immunity in rhesus macaques. Vaccine 20, 3285–3291 (2002). [DOI] [PubMed] [Google Scholar]
- 232.Arankalle VA, Chadha MS, Banerjee K, Srinivasan MA & Chobe LP Hepatitis E virus infection in pregnant rhesus monkeys. Indian J Med Res 97, 4–8 (1993). [PubMed] [Google Scholar]
- 233.Tsarev SA et al. Experimental hepatitis E in pregnant rhesus monkeys: failure to transmit hepatitis E virus (HEV) to offspring and evidence of naturally acquired antibodies to HEV. J Infect Dis 172, 31–37 (1995). [DOI] [PubMed] [Google Scholar]
- 234.Clayson ET et al. Detection of hepatitis E virus infections among domestic swine in the Kathmandu Valley of Nepal. Am J Trop Med Hyg 53, 228–232 (1995). [DOI] [PubMed] [Google Scholar]
- 235. Meng XJ et al. A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci U S A 94, 9860–9865 (1997). Hepatitis E virus was first discovered in pigs, which are now known to be the major reservoir for HEV in developed nations.
- 236.Meng XJ et al. Experimental infection of pigs with the newly identified swine hepatitis E virus (swine HEV), but not with human strains of HEV. Arch Virol 143, 1405–1415 (1998). [DOI] [PubMed] [Google Scholar]
- 237.Purcell RH et al. Pre-clinical immunogenicity and efficacy trial of a recombinant hepatitis E vaccine. Vaccine 21, 2607–2615 (2003). [DOI] [PubMed] [Google Scholar]
- 238.Usmanov RK et al. [Experimental hepatitis E infection in piglets]. Vopr Virusol 36, 212–216 (1991). [PubMed] [Google Scholar]
- 239.Arankalle VA, Chobe LP, Walimbe AM, Yergolkar PN & Jacob GP Swine HEV infection in south India and phylogenetic analysis (1985–1999). J Med Virol 69, 391–396, doi: 10.1002/jmv.10301 (2003). [DOI] [PubMed] [Google Scholar]
- 240.Cooper K. et al. Identification of genotype 3 hepatitis E virus (HEV) in serum and fecal samples from pigs in Thailand and Mexico, where genotype 1 and 2 HEV strains are prevalent in the respective human populations. J Clin Microbiol 43, 1684–1688, doi: 10.1128/JCM.43.4.1684-1688.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Kasorndorkbua C. et al. Infectious swine hepatitis E virus is present in pig manure storage facilities on United States farms, but evidence of water contamination is lacking. Appl Environ Microbiol 71, 7831–7837, doi: 10.1128/AEM.71.12.7831-7837.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Feagins AR, Opriessnig T, Huang YW, Halbur PG & Meng XJ Cross-species infection of specific-pathogen-free pigs by a genotype 4 strain of human hepatitis E virus. J Med Virol 80, 1379–1386, doi: 10.1002/jmv.21223 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Cao D. et al. Pig model mimicking chronic hepatitis E virus infection in immunocompromised patients to assess immune correlates during chronicity. Proc Natl Acad Sci U S A, doi: 10.1073/pnas.1705446114 (2017). Pigs that were iatrogenically immunosuppressed developed chronic HEV infection, and therefore are an important potential model for HEV infection in the context of immunosuppression (for example in solid organ transplant recipients).
- 244.Cossaboom CM, Cordoba L, Dryman BA & Meng XJ Hepatitis E virus in rabbits, Virginia, USA. Emerg Infect Dis 17, 2047–2049, doi: 10.3201/eid1711.110428 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Cossaboom CM et al. Cross-species infection of pigs with a novel rabbit, but not rat, strain of hepatitis E virus isolated in the United States. J Gen Virol 93, 1687–1695, doi: 10.1099/vir.0.041509-0 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Liu P. et al. Transmission of hepatitis E virus from rabbits to cynomolgus macaques. Emerg Infect Dis 19, 559–565, doi: 10.3201/eid1904.120827 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Wang L, Xia J, Wang L. & Wang Y. Experimental infection of rabbits with genotype 3 hepatitis E virus produced both chronicity and kidney injury. Gut 66, 561–562, doi: 10.1136/gutjnl-2016312023 (2017). [DOI] [PubMed] [Google Scholar]
- 248.Xia J. et al. Experimental infection of pregnant rabbits with hepatitis E virus demonstrating high mortality and vertical transmission. J Viral Hepat 22, 850–857, doi: 10.1111/jvh.12406 (2015). [DOI] [PubMed] [Google Scholar]
- 249.Huang F. et al. Experimental infection of Balb/c nude mice with Hepatitis E virus. BMC Infect Dis 9, 93, doi: 10.1186/1471-2334-9-93 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Mercer DF et al. Hepatitis C virus replication in mice with chimeric human livers. Nat Med 7, 927–933, doi: 10.1038/90968 (2001). [DOI] [PubMed] [Google Scholar]
- 251.Bissig KD et al. Human liver chimeric mice provide a model for hepatitis B and C virus infection and treatment. J Clin Invest 120, 924–930, doi: 10.1172/JCI40094 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Lutgehetmann M. et al. Humanized chimeric uPA mouse model for the study of hepatitis B and D virus interactions and preclinical drug evaluation. Hepatology 55, 685–694, doi: 10.1002/hep.24758 (2012). [DOI] [PubMed] [Google Scholar]
- 253.de Jong YP et al. Broadly neutralizing antibodies abrogate established hepatitis C virus infection. Sci Transl Med 6, 254ra129, doi: 10.1126/scitranslmed.3009512 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Winer BY et al. Recapitulation of treatment response patterns in a novel humanized mouse model for chronic hepatitis B virus infection. Virology 502, 63–72, doi: 10.1016/j.virol.2016.12.017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Carpentier A. et al. Engrafted human stem cell-derived hepatocytes establish an infectious HCV murine model. J Clin Invest 124, 4953–4964, doi: 10.1172/JCI75456 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.VanBuskirk KM et al. Preerythrocytic, live-attenuated Plasmodium falciparum vaccine candidates by design. Proc Natl Acad Sci U S A 106, 13004–13009, doi: 10.1073/pnas.0906387106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Sacci JB Jr. et al. Plasmodium falciparum infection and exoerythrocytic development in mice with chimeric human livers. Int J Parasitol 36, 353–360, doi: 10.1016/j.ijpara.2005.10.014 (2006). [DOI] [PubMed] [Google Scholar]
- 258.Mikolajczak SA et al. Plasmodium vivax liver stage development and hypnozoite persistence in human liver-chimeric mice. Cell Host Microbe 17, 526–535, doi: 10.1016/j.chom.2015.02.011 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Vaughan AM et al. Complete Plasmodium falciparum liver-stage development in liver-chimeric mice. J Clin Invest 122, 3618–3628, doi: 10.1172/JCI62684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Soulard V. et al. Plasmodium falciparum full life cycle and Plasmodium ovale liver stages in humanized mice. Nat Commun 6, 7690, doi: 10.1038/ncomms8690 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Allweiss L. et al. Human liver chimeric mice as a new model of chronic hepatitis E virus infection and preclinical drug evaluation. J Hepatol 64, 1033–1040, doi: 10.1016/j.jhep.2016.01.011 (2016). [DOI] [PubMed] [Google Scholar]
- 262.Sayed IM et al. Transmission of hepatitis E virus infection to human-liver chimeric FRG mice using patient plasma. Antiviral Res 141, 150–154, doi: 10.1016/j.antiviral.2017.02.011 (2017). [DOI] [PubMed] [Google Scholar]
- 263.Sayed IM et al. Study of hepatitis E virus infection of genotype 1 and 3 in mice with humanised liver. Gut 66, 920–929, doi: 10.1136/gutjnl-2015-311109 (2017). [DOI] [PubMed] [Google Scholar]
- 264. van de Garde MD et al. Hepatitis E Virus (HEV) Genotype 3 Infection of Human Liver Chimeric Mice as a Model for Chronic HEV Infection. J Virol 90, 4394–4401, doi: 10.1128/JVI.00114-16 (2016). Liver chimeric mice were able to support chronic infection with HEV genotype 3, establishing a murine model to study the virus.
- 265.Washburn ML et al. A humanized mouse model to study hepatitis C virus infection, immune response, and liver disease. Gastroenterology 140, 1334–1344, doi: 10.1053/j.gastro.2011.01.001 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Bility MT et al. Hepatitis B virus infection and immunopathogenesis in a humanized mouse model: induction of human-specific liver fibrosis and M2-like macrophages. PLoS Pathog 10, e1004032, doi: 10.1371/journal.ppat.1004032 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Billerbeck E. et al. Humanized mice efficiently engrafted with fetal hepatoblasts and syngeneic immune cells develop human monocytes and NK cells. J Hepatol 65, 334–343, doi: 10.1016/j.jhep.2016.04.022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Kabrane-Lazizi Y. et al. Evidence for widespread infection of wild rats with hepatitis E virus in the United States. Am J Trop Med Hyg 61, 331–335 (1999). [DOI] [PubMed] [Google Scholar]
- 269. Debing Y. et al. A rat model for hepatitis E virus. Dis Model Mech 9, 1203–1210, doi: 10.1242/dmm.024406 (2016). Rats were infected with a rat-human chimeric HEV strain and were responsive to antiviral therapy, suggesting they could be a potential animal model for HEV studies.
- 270.Purcell RH et al. Hepatitis E virus in rats, Los Angeles, California, USA. Emerg Infect Dis 17, 2216–2222, doi: 10.3201/eid1712.110482 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Hong Y. et al. Experimental infection of Z:ZCLA Mongolian gerbils with human hepatitis E virus. World J Gastroenterol 21, 862–867, doi: 10.3748/wjg.v21.i3.862 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Li W. et al. Experimental infection of Mongolian gerbils by a genotype 4 strain of swine hepatitis E virus. J Med Virol 81, 1591–1596, doi: 10.1002/jmv.21573 (2009). [DOI] [PubMed] [Google Scholar]
- 273.Yang Y. et al. Effect of swine hepatitis E virus on the livers of experimentally infected Mongolian gerbils by swine hepatitis E virus. Virus Res 208, 171–179, doi: 10.1016/j.virusres.2015.06.007 (2015). [DOI] [PubMed] [Google Scholar]