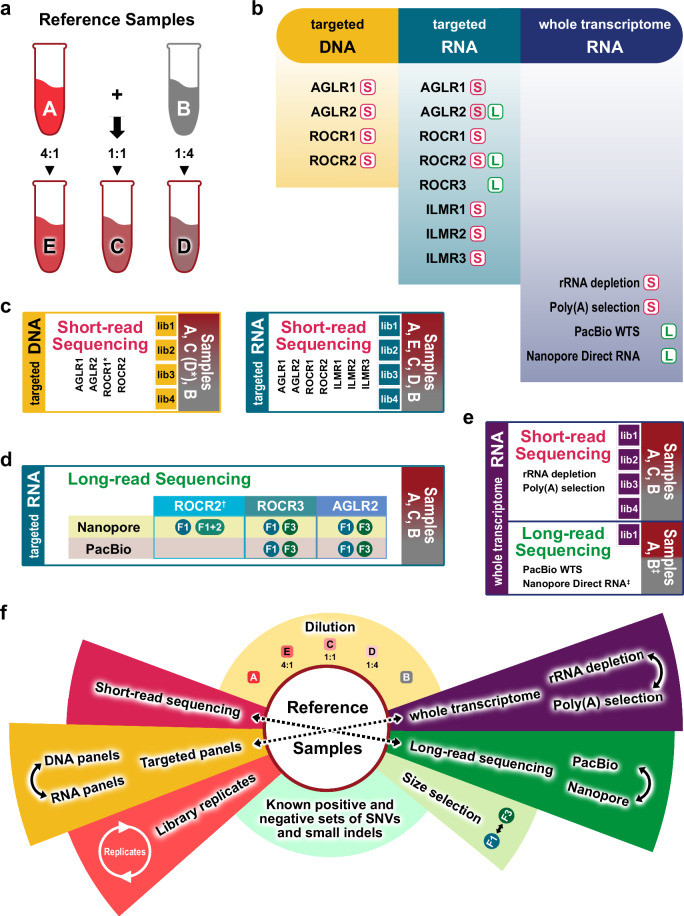
Fig. 1.
Illustration of study design. (a) To create reference samples C, D, and E: samples A and B were mixed in the ratios of 1:1, 1:4, and 4:1 respectively. (b) Three types of libraries were prepared for the reference samples: targeted RNA, targeted DNA, and whole transcriptome RNA. The libraries were sequenced using short-read or long-read sequencing methods, or both. The panel codes are explained in Table 2. The pink “S” represents short-read sequencing, while green “L” represents long-read sequencing. The ILMR3 panel is a whole exome RNA panel, and it was placed under “targeted RNA” for visual simplicity. (c) Both targeted DNA and targeted RNA libraries were sequenced with short-read sequencing. For targeted DNA libraries, four library replicates (lib1-4) were prepared for Samples A, B, and C using AGLR1, AGLR2, and ROCR2. * Three library replicates (lib1-3) were prepared using ROCR1. Sample D was sequenced with ROCR1 instead of Sample C. For targeted RNA libraries, four library replicates (lib1-4) were prepared for Samples A, B, C, D, and E using 7 panels. (d) Targeted RNA libraries of Sample A, B, and C were made with three panels, each library was split into different fractions (F1, F1 + 2, or F3), and sequenced with both long-read sequencing platforms. (e) † ROCR2 was only used for Sample A and the libraries was sequenced only on Nanopore. ‡ Sample B was sequenced by Nanopore Direct RNA protocol only. (f) An illustration shows the flexible options for possible comparison analyses for an in-depth study of the impacts of targeting, size selection, and sequencing protocols.