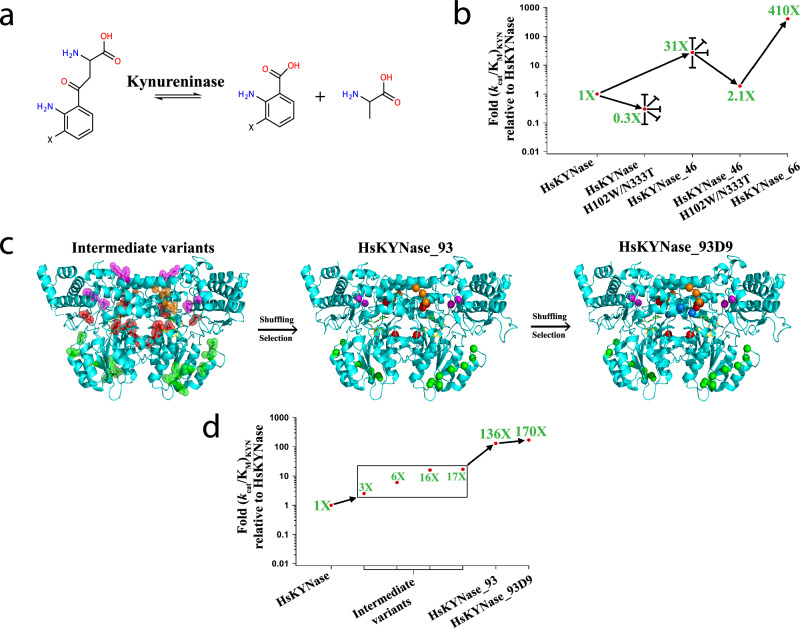
Fig. 1. Evolutionary trajectories that led to the identification of generalist HsKYNase_93D9 and specialist HsKYNase_66.
a Reaction(s) catalyzed by Kynureninases (X denotes either -H or -OH groups). b Schematic showing the directed evolution trajectory leading to the isolation of HsKYNase_6623. Blunted arrows denote dead-end evolutionary intermediates, from which higher-catalytic-activity variants could not be selected from numerous libraries. c Structural representation of the mutations in key intermediates in the evolutionary trajectory to HsKYNase_93D; Intermediate variants comprising HsKYNase_1 (mutated amino acids in magenta), HsKYNase_8 (orange), high B-factor_10 (green) and rationally designed variants (red) were shuffled and then selection led to the isolation of HsKYNase_93 (for more details see main text, Supplementary Fig. 2, Supplementary Tables 1–7). Center: Location of the 15 amino acid substitutions of HsKYNase_93 is shown in spheres color-coded as described above. Right: Mutations in HsKYNase_93D9 showing in blue the two additional substitutions located in a critical active site loop (see Supplementary Fig. 2 and Supplementary Tables 1–7). The PLP co-factor and the 3-hydroxy-hippuric acid inhibitor are shown in yellow and green sticks respectively. The inhibitor is color-coded by atom (N: blue, O: red). d Schematic summarizing the directed evolution trajectory leading to the isolation of HsKYNase_93D9. Combination of mutations found in intermediate variants such as HsKYNase_1 and HsKYNase_8 (both showing KYN improvement of ~17-fold) with those from high B-factor_10 (6-fold) and rationally designed variants (3- and 16-fold) (Table 2 and Supplementary Tables 3, 4) led to the identification of HsKYNase_93 and finally HsKYNase_93D9.