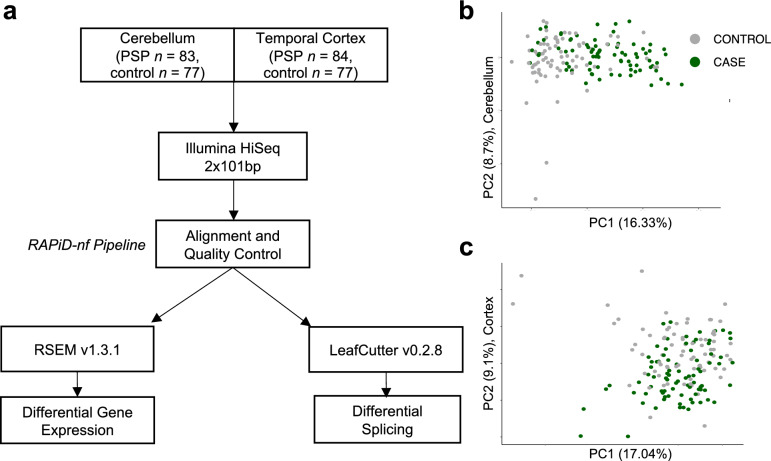
Fig. 1.
RNA sequencing pipeline using RAPiD-nf aligns and quality controls 161 samples for differential expression and splicing analysis. a RNA-seq samples were obtained from Synapse.org [50]. The cohort included 161 PSP cases and controls, with samples taken from the cerebellum and temporal cortex. Samples were processed using RAPiD-nf, a processing pipeline in the NextFlow framework. RAPiD-nf uses Trimmomatic, STAR, FASTQC, featureCounts, Pathogen, and Picard for pre-processing and quality control. RSEM and Limma were used for differential gene expression, and Regtools and LeafCutter were used for differential intron excision. b, c Principal component analysis of the RNA-seq expression matrix after covariate adjustment in cerebellum and temporal cortex shows no remaining outliers. 161 samples were included in downstream analysis