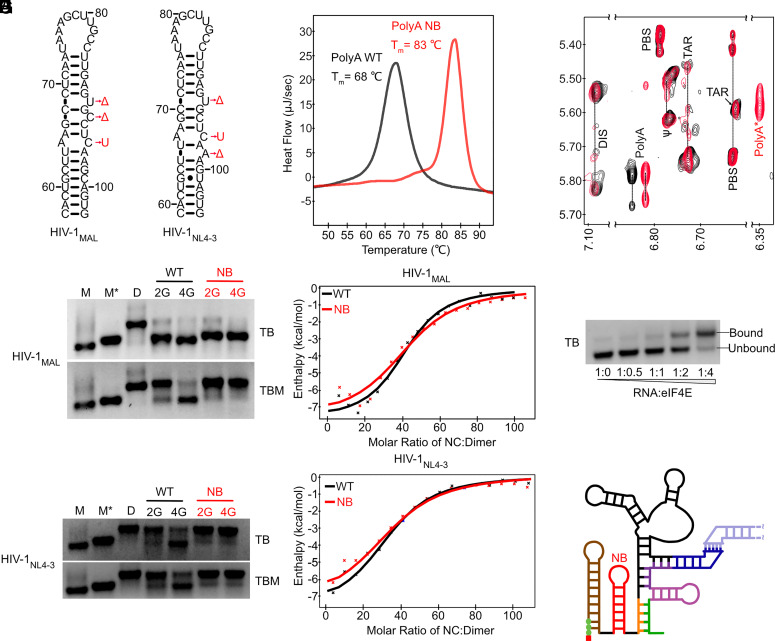
Fig. 2.
polyANB-modified 3G RNAs form cap-exposed dimers. (A) NMR-derived 5′ polyA secondary structures [HIV-1MAL (9) and HIV-1NL4-3 (12)] and polyA-stabilizing mutations (polyANB; Δ = deletion, U = uracil substitution. (B) Increased melting temperature of the HIV-1MAL polyANB hairpin (+15 °C; measured by DSC). (C and D) In vitro dimerization assays for wild-type (WT) and polyANB-mutated (NB) HIV-1MAL and HIV-1NL4-3 leader RNAs performed with (TBM) and without (TB) 0.2 mM MgCl2 in the gel and running buffer. Controls showing the monomer (M), monomer in dimeric conformation (M*) (9), and dimer (D) bands. (E and F) HIV-1MAL (E) and HIVNL4-3 (F) leader RNAs exhibit similar NC binding properties by ITC. (G) Regions of NOESY spectra obtained for HIV-1MAL-2G (black) and HIV-1MAL 4G polyANB (red) leader RNAs prepared with nucleotide-specific 2H-labeling (A2GrUr; protons only at adenosine C-2 and ribose and guanosine/uridine ribose positions). Assignments are from ref. 9. polyA* denotes shifted signal due to polyANB mutations. (H and I) Dimeric HIV-1MAL Cap3G polyANB binds eIF4E (H), consistent with an exposed 5′ cap (I).