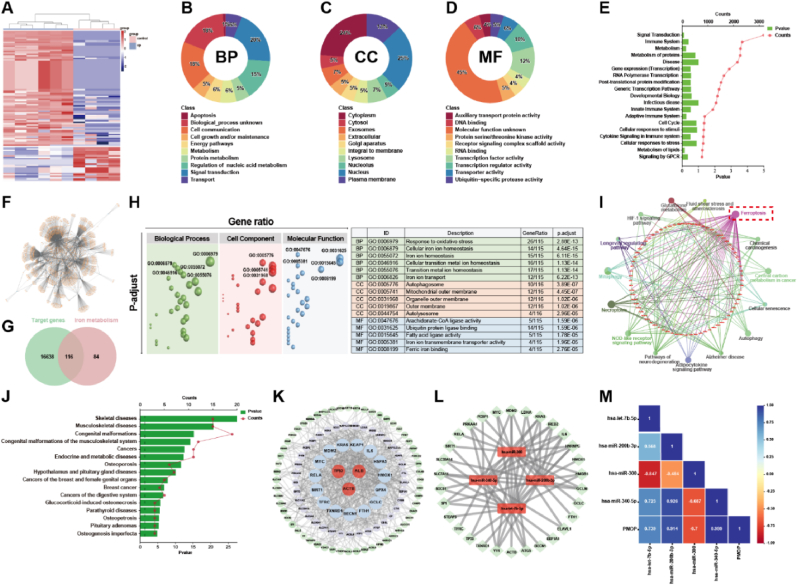
Fig. 1.
(A) The differentially expressed miRNAs based on the plasma samples of postmenopausal patients with (Group B) and without (Group A) osteoporosis; (B) Biological processes, (C) Cellular component, and (D) Molecular function based on the differentially expressed miRNA; (E) The key reactome pathways of the differentially expressed miRNAs; (F) Graph of the corresponding target gene network of the differentially expressed miRNA; (G) The Venn diagram of target gene set and iron metabolism gene set; (H) GO enrichment results of the union 116 genes; (I) KEGG pathways of the union 116 genes; (J) Disease analysis of the union 116 genes; (K) Protein-protein interaction network and the hub genes of the union 116 genes; (L) The hub miRNAs and their correlation with the top 50 hub genes; (M) Pearson correlation coefficient between the hub miRNAs and the occurrence of PMOP. │log2FC│>2, P-value <0.05.