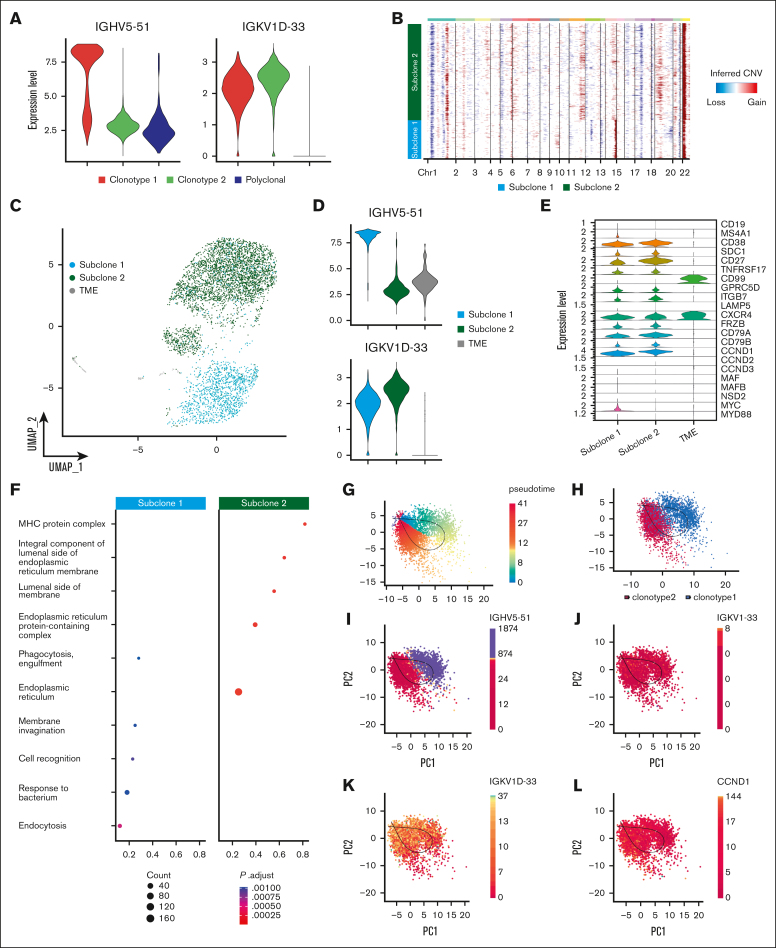
Figure 4.
Heavy-chain locus loss characterization. (A) Violin plot of 2 MM BCR clonotypes of sample P1, representing the expression of heavy and light chains in each clonotype. Clonotype 1 is represented in red, clonotype 2 in green, and polyclonal cells in blue. (B) CNAs inferred by inferCNV pipeline, to define functional differences between the 2 tumor clonotypes. Subclone 1 is represented in light blue and subclone 2 in green. (C) Color coded map of subclones distribution in the UMAP space, with the same color code of panel B. Tumor microenvironment (TME) is shown in gray. (D) Violin plot representing the expression of heavy and light chain in each subclone. (E) Analysis of marker genes by subclone is represented by violin plots. (F) Pathway enrichment analysis, comparing the transcriptional profiles of CNAs subclones, (.1 as P value cutoff; Padjusted (Padj) method by Benjamini-Hochberg; see “Materials and Methods”). (G-K) After normalization and projection on a reduced-dimensional space (using UMAP), the cells were colored by pseudotime inference (G), clonotype origin (H), IGHV5-51 gene expression (I), IGKV1-33 gene expression (J), IGKV1D-33 gene expression (K) and CCND1 gene expression (L). Each model is fitted using the trajectory inferred by slingshot.