Summary
Tomato (Solanum lycopersicum) stands as one of the most valuable vegetable crops globally, and fruit firmness significantly impacts storage and transportation. To identify genes governing tomato firmness, we scrutinized the firmness of 266 accessions from core collections. Our study pinpointed an ethylene receptor gene, SlEIN4 , located on chromosome 4 through a genome‐wide association study (GWAS) of fruit firmness in the 266 tomato core accessions. A single‐nucleotide polymorphism (SNP) (A → G) of SlEIN4 distinguished lower (AA) and higher (GG) fruit firmness genotypes. Through experiments, we observed that overexpression of SlEIN4 AA significantly delayed tomato fruit ripening and dramatically reduced fruit firmness at the red ripe stage compared with the control. Conversely, gene editing of SlEIN4 AA with CRISPR/Cas9 notably accelerated fruit ripening and significantly increased fruit firmness at the red ripe stage compared with the control. Further investigations revealed that fruit firmness is associated with alterations in the microstructure of the fruit pericarp. Additionally, SlEIN4 AA positively regulates pectinase activity. The transient transformation assay verified that the SNP (A → G) on SlEIN4 caused different genetic effects, as overexpression of SlEIN4 GG increased fruit firmness. Moreover, SlEIN4 exerts a negative regulatory role in tomato ripening by impacting ethylene evolution through the abundant expression of ethylene pathway regulatory genes. This study presents the first evidence of the role of ethylene receptor genes in regulating fruit firmness. These significant findings will facilitate the effective utilization of firmness and ripening traits in tomato improvement, offering promising opportunities for enhancing tomato storage and transportation capabilities.
Keywords: Tomato, EIN4, single‐nucleotide polymorphism, fruit firmness, fruit pericarp microstructure, pectinase activity
Introduction
Tomato (Solanum lycopersicum) is a high‐value crop with worldwide markets and uses (Vincent et al., 2013). The tomato fruit industry faces a critical hurdle in postharvest deterioration, contributing to 50% of harvest losses (Thole et al., 2020). The firmness of tomato fruits has been identified as a critical characteristic for determining fruit maturity and storability. The decline in fruit firmness is because of the breakdown of cell wall polymers by cell wall hydrolases (Su et al., 2022). Currently, enzymes such as polygalacturonase (PG), pectin lyase (PL), pectinesterase (PE), pectin methylesterase (PME) and gibberellin 2‐oxidase (GA2ox) have been determined to have a role in fruit softening. PG exhibits the highest activity among these cellular hydrolases and was the first to be discovered. Employing antisense down‐regulation of PG has effectively diminished softening and water loss in apples and strawberries (Atkinson et al., 2012; Shi et al., 2021). Silencing of the PL gene in tomatoes results in harder fruit without a significant effect on ripening (Brummell and Harpster, 2001; Wang et al., 2019). PE has been reported in many plant tissues and implicated in many developmental processes, including cellular adhesion, stem elongation (Micheli, 2001) and fruit ripening (Brummell and Harpster, 2001). In addition, studies have shown that PG‐mediated pectin degradation is enhanced by PME‐mediated pectin demethylation (Wakabayashi et al., 2003). A mutation in the tomato FIRM SKIN 1 (FIS1) gene, which encodes a GA2 oxidase, has been shown to enhance the biosynthesis of cutin and wax, thereby increasing the firmness of the fruit (Li et al., 2020). Several quantitative trait loci (QTLs) linked to firmness traits have been confirmed in tomatoes (Alexander and Grierson, 2002; Ma et al., 2014). For instance, six QTLs located on chromosomes 1, 3, 4, 6, 9 and 11 have been determined in the Peruvian tomato Lycopersicon parviflorum1706 (LA1706) (Fulton et al., 1997), as well as three QTLs for increasing fruit firmness which were identified in the Pannelli tomato LA1657 (Frary et al., 2004). A QTL for fruit firmness was mapped to tomato chromosome 2 using the Zamir Solanum pennellii interspecific introgression lines (ILs) and fine‐mapped in a population consisting of 7500 F2 and F3 lines from IL 2–3 and IL 2–4. This firmness QTL contained five distinct subpeaks, Firs.p.QTL2.1 to Firs.p.QTL2.5. An ethylene response factor (ERF) underlying Firs.p.QTL2.2 and a region containing three PME genes underlying Firs.p.QTL2.5 were nominated as QTL candidate genes (Chapman et al., 2012).
Fruit firmness is tightly linked to fruit ripening (Hu et al., 2020; Monsalve et al., 2022; Usenik et al., 2008). It has been documented that ethylene plays a critical role in regulating the ripening process of fruit (Huang et al., 2022; Oetiker and Yang, 1995). The ethylene production commences with methionine, which undergoes a transition to S‐adenosine‐L‐methionine. Subsequently, it transforms into 1‐aminocyclopropane‐1‐carboxylic acid (ACC). Furthermore, ACC undergoes oxidation, leading to the formation of ethylene, cyanide and carbon dioxide (Wang et al., 2002; Yang and Hoffman, 1984). Several plant species have well‐established signal transduction pathways involving ethylene (Gapper et al., 2013; Hua and Meyerowitz, 1998). Based on earlier research, the perception of the ethylene signal involves ETHYLENE RESPONSE1 (ETR1), ETR2, ETHYLENE RESPONSE SENSOR1 (ERS1), ERS2 and ETHYLENE INSENSITIVE4 (EIN4) in Arabidopsis (Adams‐Phillips et al., 2004; Alonso et al., 1999; Stepanova and Ecker, 2000). Tomato exhibits seven ethylene receptor group representatives, including SlETR1, SlETR2, SlETR3 (NEVER‐RIPE), SlETR4, SlETR5, SlETR6 and SlETR7. According to protein structure classification, SlETRl‐3 belongs to subfamily 1, while SlETR4‐7 belongs to subfamily 2. All these members have a vital part to play in the fruit ripening process (Chen et al., 2020; Kevany et al., 2007, 2008; Mubarok et al., 2019; Wilkinson et al., 1995). SlETR1 and SlETR2 are consistent with the AtETR1 sequence structure of Arabidopsis. SlETR3 and AtERS1 have similar sequences without response regulatory domains. The SlETR4‐7 structure is similar to AtEIN4 and AtETR2 (Chen et al., 2020; Klee and Tieman, 2002; Tieman et al., 2001). In Arabidopsis, EIN4 belongs to the ETR2 subfamily, and its absence, along with ETR1, significantly diminishes the reaction of Arabidopsis to ethylene (Liu and Wen, 2012). Few studies of EIN4 have been reported on tomatoes so far. Despite this crucial role, most studies have primarily concentrated on investigating the impact of ethylene receptors on ethylene response and fruit ripening, with only a limited amount of reports addressing the direct regulation of fruit firmness by ethylene receptors.
The advent of genome‐wide association studies (GWAS) has revolutionized the investigation of complex traits and locus interactions in plants, offering a potent approach to unravelling the genetic underpinnings of these phenomena (Nordborg and Weigel, 2008). In our research, we conducted GWAS to explore fruit firmness traits across 266 diverse tomato germplasms. Through this study, we successfully identified an ethylene receptor, SlEIN4, located on chromosome 4, which exhibited a single‐nucleotide polymorphism (SNP) (A → G) distinguishing lower (AA) and higher (GG) fruit firmness materials. Further experimentation revealed that the overexpression of SlEIN4 (AA) significantly delayed tomato fruit ripening and reduced fruit firmness considerably. The SNP (A → G) on SlEIN4 causes different genetic effects on fruit firmness. Instead, gene editing of SlEIN4 using CRISPR/Cas9 noticeably accelerated fruit ripening, and markedly increased fruit firmness compared with the control. Our findings first reported that SlEIN4 influences fruit firmness and ripening, promoting the effective use of firmness and ripening traits in tomato improvement.
Results
Sequencing, variants and population structure of natural tomato populations
For this investigation, we employed 266 tomato resources for genotyping, followed by GWAS analysis. The resources used for this study (listed in Table S1) comprised 149 large tomatoes (S. l. lycopersicum, BIG), 103 cherry tomatoes (var cerasiforme, CER) and 14 currant tomatoes (Solanum pimpinellifolium, PIM), all of them were genotyped in a previous study (Lin et al., 2014). Cluster analysis was conducted using genome‐wide SNPs on 266 tomato materials to uncover tomato population structure. We genotyped these materials using 5 485 483 high‐quality SNPs, and the heterozygosity of the two SNP markers fell within the acceptable range (<25%). Consistent with the previous report, these materials were separated into three groups (Figure S1a), further supporting our group structure analysis (Figure S1b,c). We conducted principal components analysis (PCA) and model‐based cluster analysis based on whole‐genome SNPs for BIG, CER and PIM accessions to determine the genetic changes across the three groups (Figure S1b–d). The largest principal component (PC1) was responsible for 70.53% of the domestication‐related variation, and the second component (PC2) accounted for 18.47% of the variation. By comparison, the third component (PC3) elucidated 11% of the variation (Figure S1c). Our admixture analysis further confirmed the genetic structure (Figure S1b). Population structure and PCA upheld the three biological classifications. However, BIG and CER could not be distinguished entirely due to their complicated genetic background.
GWAS identification of SlEIN4 as a candidate gene for firmness determination
The experimental population was cultivated at two locations: open‐field cultivation at Huazhong Agricultural University (HZ) and greenhouse cultivation at Zhongdou Seed Company (ZD). Initially, we analysed the survey data of tomato fruit firmness. We observed good repeatability in the results of the two replicates. The fruit firmness values of the 266 natural populations ranged from 3 to 12 kg/cm2 (Table S1), with a coefficient of variation of 21.8% among the population materials. Fruit firmness remained stable across the two experiment locations (R = 0.44), and the trait exhibited a normal distribution within the panel of accessions (Figure 1a,b). We employed re‐sequencing technology to screen 5.5 million high‐quality SNPs. The GWAS analysis on the determined fruit firmness values was conducted using the genome‐wide efficient mixed‐model association (GEMMA) model. We identified 50 sites that showed significant associations with fruit firmness (Table S2). During GWAS, several significant SNPs linked to fruit firmness were identified, and no inflation was observed throughout the P‐values distributed at the optimal step of the model (Figure 1c,d). This indicates that the population structure was effectively controlled in the study. Five clear peaks were distributed across chromosomes 4, 5, 7, 9 and 11, with the peak value on chromosome 4 being the highest (Figure 1c). Therefore, we focused on the most significant peak SNP site ch04_55155437 (release SL2.40) on chromosome 4 (P‐value = 4.73 × 10−11).
Figure 1.
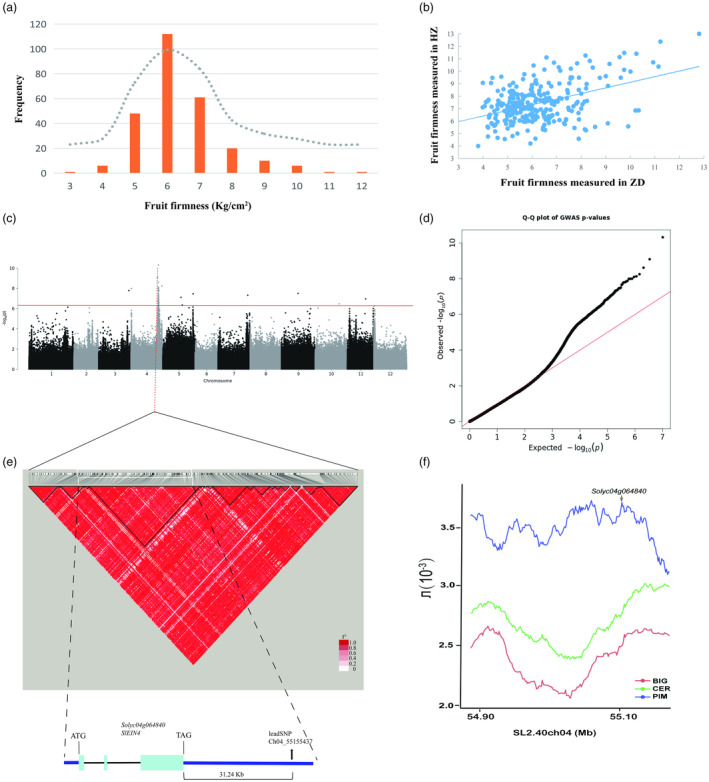
Fruit firmness correlation analysis results. (a) Distribution of adjusted fruit firmness values in the collection of 266 tomato samples. (b) Correlation between fruit firmness values at two different sampling sites from the 266 materials. (c) Manhattan chart displaying the genome‐wide association study (GWAS) results of fruit firmness (MLM, N = 266). The y‐axis values represent the negative logarithm of the association P‐value, with the significance threshold indicated by a horizontal line (SL2.40 version). (d) Quantile‐quantile plot for fruit firmness in the GWAS population. (e) Pairwise r2 values (a measure of LD) among the polymorphic sites of ch04_55155437 (release SL2.40). (f) Genomic distribution of PIM, CER and BIG nucleotide diversity in domestication sweeps containing SlEIN4 on chromosome 4 (SL2.40 version).
In our subsequent analysis, the pairwise linkage disequilibrium (LD) estimates (rs2) for the genomic location were utilized to narrow down the genomic interval and search for potential candidate genes near the peak SNP (Figure 1e). We determined the physical positions of the SNPs using the tomato genomic sequence, version SL2.40, which can be obtained from the website (http://solgenomics.net/). By conducting a genetic linkage analysis of SNP locus ch04_55155437 (with a P‐value = 4.73 × 10−11), we obtained a local view of the extent of LD decay within the region of 55 127 814–55 157 228. Within this region, an ethylene receptor, a MADS‐box transcription factor, a transmembrane protein 34 gene, and two pathogenesis‐related proteins were contained (Table S3). Among them, only the ethylene receptor showed a non‐synonymous SNP mutation. Through gene expression profiling on the website (http://ted.bti.cornell.edu/), it was discovered that the ethylene receptor was expressed explicitly in fruit, the MADS‐box transcription factor was expressed in all tissues, while others showed minimal expression in fruit. Notably, the π values indicated a significant reduction in the ethylene receptor interval for the BIG and CER groups compared with the PIM group (Figure 1f), providing evidence that it was indeed under selection. By blasting the sequence in NCBI (https://www.ncbi.nlm.nih.gov/), it was shown to be an EIN4‐like ethylene receptor with a histidine kinase domain in the amino acid sequence (Figure S2), and we named it SlEIN4.
Identification of a critical base mutation in SlEIN4 tightly associated with tomato fruit firmness
The genetic structure of SlEIN4 was thoroughly analysed through Gene Structure Display Server, GSDS (http://gsds.cbi.pku.edu.cn/), revealing a deoxyribonucleic acid (DNA) length of 2841 bp consisting of three exons and two introns. Its cDNA length was 546 bp, encoding a protein of 181 amino acids (Figures S2 and S3). Amino acid sequence alignment of SlEIN4 and SlETR1‐SlETR7 evolutionary tree analysis showed that SlEIN4 and SlETR7 had relatively higher homology (Figures S4 and S5). During our investigation, a base mutation (A‐G) was detected in the third exon of the SlEIN4 DNA sequence among the 266 materials (release SL2.40) (Figure 2a, Table S1). Haplotype AA was predominantly detected in the BIG group, while haplotype GG was exclusively found in the PIM group (Figure 2b). This mutation led to an amino acid change in the sequence from lysine (Lys) to glutamate (Glu). Among the 35 high‐firmness varieties with firmness greater than 8 kg/cm2, haplotype GG accounted for 60.000%, haplotype AA accounted for 37.143%, and AG accounted for 2.857%. Conversely, in the 231 low‐firmness varieties with firmness less than or equal to 8 kg/cm2, haplotype AA accounted for 77.056%, haplotype GG accounted for 17.316%, and AG accounted for 5.628%. The gene sequencing results also revealed that all the haplotype GG was identified in the top 15 high‐firmness cultivars. By comparison, all the haplotype AA was identified in the top 15 low‐firmness cultivars within the natural population (Figure 2c). Among the 252 homozygous genotypes of SlEIN4 in the natural population, two major haplotypes, on account of the leader SNP (SL2.40ch04_55126611), were relevant to high‐firmness and low‐firmness properties observed in tomatoes, respectively. The SNP, accompanied by the most elevated correlation with fruit firmness, accounted for 15.36% of the total variance (Figure 2d). According to these compelling findings, SlEIN4 emerged as a prime candidate gene for determining fruit firmness.
Figure 2.
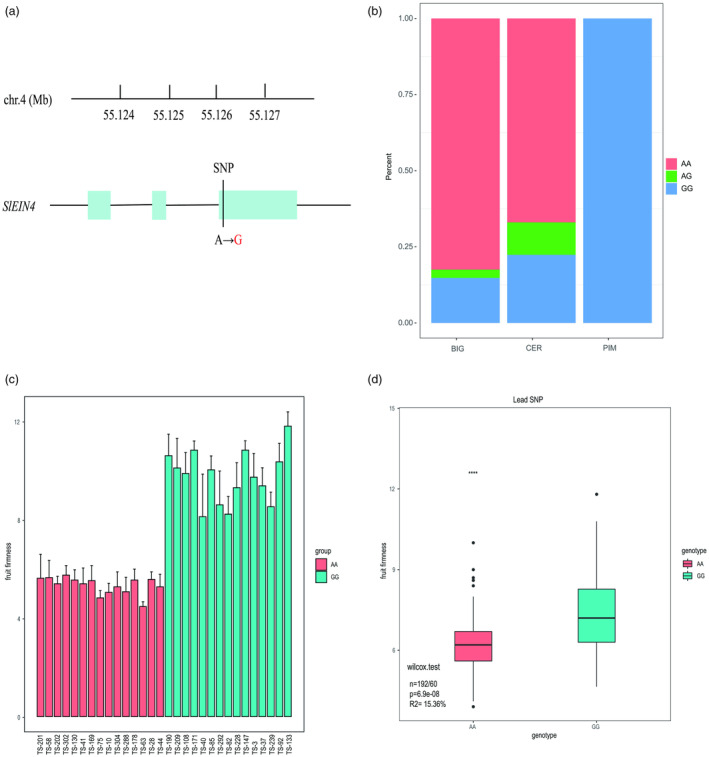
Lead single‐nucleotide polymorphism (SNP) in SlEIN4. (a) The gene structure of the ethylene receptor (SlEIN4) is strongly associated with an SNP in the third exon, with genomic coordinates from SL2.40. (b) Distribution of strongly associated SNPEIN4 among the PIM, CER and BIG groups. (c) The SNPs of the SlEIN4 gene in 15 high‐fruit firmness and 15 low‐fruit firmness cultivars. (d) Box plot of stomatal densities in tomato accessions with different alleles (A or G) at SNP ch04_55126611. For each box plot, the horizontal line in the box indicates the median value, the box height indicates the 25th to 75th percentiles of the total data, the whiskers indicate the interquartile range, and the outer dots indicate outliers.
SlEIN4 was primarily expressed in tomato fruit
We delved into its subcellular localization and expression pattern for a more comprehensive understanding of SlEIN4's function. Our findings revealed that SlEIN4 was situated on the cell membrane, indicating it functions as a membrane protein (Figure 3a). Through quantitative RT‐PCR (RT‐qPCR) analysis of different tissues, including root, stem, leaf, flower and different fruit stages, we observed notably higher expression levels of SlEIN4 in fruit compared with other tissues, with the highest expression in mature green fruits (Figure 3b). To further explore its expression in situ, we generated transgenic tomatoes overexpressing the glucuronidase synthase (GUS) gene under the control of the SlEIN4 promoter. GUS staining suggested that the SlEIN4 gene is expressed in the leaf bud, fruits at the mature green (MG) stage, breaker (BR) stage, yellow ripening (YR) stage and red ripening (RR) stage (Figure 3c). Notably, the GUS staining was significantly darker in fruit than in other tissues, suggesting that the SlEIN4 gene primarily plays a critical role in fruit development.
Figure 3.
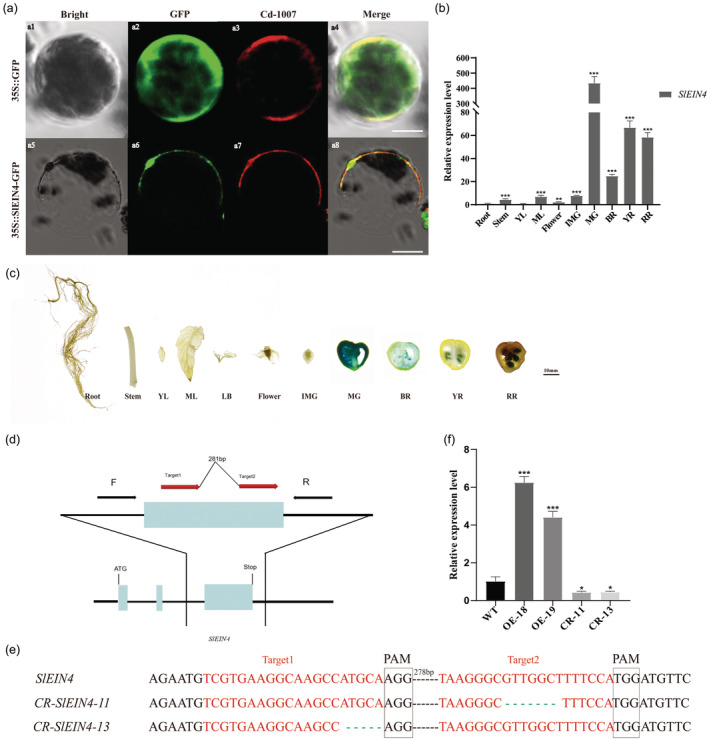
Subcellular localization and expression of the SlEIN4 gene and characterization of overexpression and CRISPR/Cas9‐engineered mutation lines. (a) The subcellular localization of SlEIN4 in tobacco protoplast. (a‐1 to a‐4) Fluorescence signals derived from 35 S::GFP and mCherry in tobacco protoplast. (a‐5 to a‐8) Fluorescence signals derived from 35 S::SlEIN4‐GFP and mCherry in tobacco protoplast. Scale bars = 20 μm. (b) Relative expression of SlEIN4 in different Micro Tom (MT) tissues by qPCR. Root, roots; Stem, stems; YL, young leaves; ML, mature leaves; Flower, flower; IMG, immature green fruit; MG, mature green fruit; BR, breaker fruit; YR, yellow ripening fruit; RR, red ripening fruit. The quantitative fluorescence results are shown from three biological and three technical replicates. The Actin gene (Solyc11g005330) was used as the internal control. Data are presented as means ± SD (n = 3). Asterisks indicate the significant difference between root and other tissues revealed by t‐test: *P < 0.05, **P < 0.01, ***P < 0.001. (c) The SlEIN4 promoter drives GUS gene expression in MT. Tissues of transgenic tomato plants were stained with X‐Gluc. Root, roots; Stem, stems; YL, young leaves; ML, mature leaves; LB, leaf bud; Flower, flower; IMG, immature green fruit; MG, mature green fruit; BR, breaker fruit; YR, yellow ripening fruit; RR, red ripening fruit. Scale bars = 10 mm. (d) Schematic illustration of the two sgRNA target sites (red arrows) in SlEIN4. Black arrows represent the locations of PCR genotyping primers. (e) CR‐SlEIN4 alleles were identified from two T0 mutant lines. Allele sequences, as determined by sequencing, are shown. Both CR‐SlEIN4‐11 and CR‐SlEIN4‐13 had base deletions near the sgRNA target site. Red text indicates the sgRNA target sequence, and black boxes indicate protospacer‐adjacent motif (PAM) sequences. (f) The relative expression level of SlEIN4. Values are presented as means ± SD (n = 3). Asterisks indicate the significant difference between wild‐type and transgenic plants revealed by t‐test: *P < 0.05, **P < 0.01, ***P < 0.001.
To examine the role of SlEIN4 in fruit development, we generated both SlEIN4 (AA) gene editing lines and overexpression lines. For this purpose, Micro Tom (MT) was used as the genetic background material due to its low firmness and haplotype AA of SlEIN4. Two targets for the CRISPR/Cas9 recombinant vector were identified in Figure 3d (See primers in Table S4). Subsequently, two gene editing lines of SlEIN4, namely CR‐11 and CR‐13, were identified through gene sequencing (Figures S6 and S7). CR‐11 exhibited a 7‐bp deletion near the second sgRNA target, while CR‐13 showed a 5‐bp deletion near the first sgRNA target in SlEIN4 (Figure 3e). Furthermore, two overexpression lines, OE‐18 and OE‐19, with elevated expression levels of SlEIN4, were chosen for additional analysis (Figure 3f). Interestingly, we observed a distinct decrease in the expression of SlEIN4 in the two gene‐edited lines (Figure 3f).
SlEIN4 negatively regulates tomato fruit ripening by affecting ethylene downstream signalling
Monitoring the fruit development of transgenic and wild‐type (WT) plants from 27 days after flowering, the results revealed no notable difference in appearance between WT and OE lines (Figure 4a,b). Moreover, the ripening of CR lines was remarkably accelerated, occurring probably 2–3 days earlier than WT. Conversely, the ripening of OE lines was dramatically delayed by around 3 days compared with the control (Figures 4a,c and S8). The ethylene release and ACC content of MT, CR and OE transgenic materials were detected at the same time on Days 31, 34, 37, 39 and 41 after flowering, respectively. The ethylene release of CR is earlier than that of MT, while the ethylene release of OE is later than that of MT (Figure 4d). As a precursor of ethylene synthesis, ACC content in fruit directly determines the level of ethylene release. It was found that the peak of ACC content in CR appears the earliest, while OE appears the latest among WT and transgenic lines (Figure 4e). These data were consistent with the results of ethylene production. Considering the functional redundancy of ethylene receptors, as well as the unclear relative contribution of each receptor to ethylene signalling (Chen et al., 2020), we investigated the expression levels of several genes associated with ethylene signal pathways, including Constitutive Triple Response1 (CTR1), Ethylene Insensitive 2 (EIN2), ETR5 and EIN3, in mature green fruits. The expression of CTR1 was dramatically increased in OE (Figure 4f), while the transcription of both ETR5 and EIN2 was significantly increased in CR (Figure 4g,i). However, the mRNA level of EIN3 was significantly increased in CR lines, whereas it was dramatically decreased in OE lines (Figure 4h).
Figure 4.
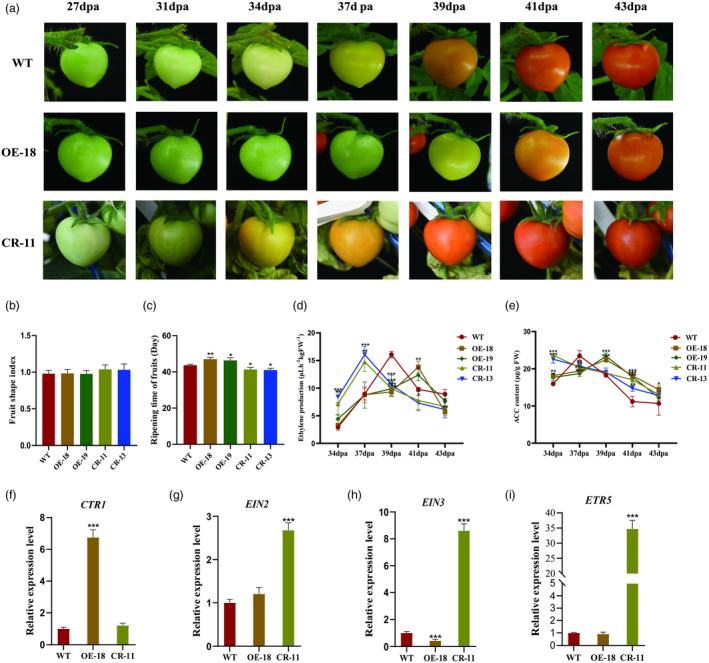
Fruit ripening time, shape index, 1‐aminocyclopropane‐1‐carboxylic acid (ACC) content, and ethylene production of wild‐type (WT) and transgenic lines. (a) Representative fruits of WT, OE‐18 and CR‐11 from 27 dpa to 43 dpa. (b‐c) Fruit shape index and ripening time of WT and transgenic lines. (d‐e) ethylene production and ACC content of WT and transgenic fruits at 34 dpa, 37 dpa, 39 dpa, 41 dpa and 43 dpa. (f‐i) The relative expression levels of CTR1, EIN2, EIN3 and ETR5 in WT, OE‐18 and CR‐11 fruits at the mature green fruit (MG) stage. Values are presented as means ± SD (n = 3). Asterisks indicate the significant difference between WT and transgenic plants revealed by t‐test: *P < 0.05, **P < 0.01, ***P < 0.001.
SlEIN4 negatively regulates tomato firmness by affecting the fruit pericarp microstructure and pectinase activity
To uncover the role of SlEIN4 on fruit firmness determination, we measured the fruit firmness across different stages in WT, OE and CR lines. At the BR and YR stage, the fruit firmness of both WT, OE and CR lines remained high, with no significant difference (Figure 5a,b). Notably, the fruit firmness of OE lines was dramatically lower than the control, while it was markedly higher in CR lines compared with the control at the RR stage (Figure 5c). These results indicate that SlEIN4 negatively regulates tomato fruit firmness. We also assessed the fruit weight and quality in the RR stage of WT and transgenic lines. The fruit weight and sugar‐acid ratio showed no apparent difference between WT and transgenic lines (Figure S9b,c). However, the total soluble solids in CR fruits were notably lower than in WT and OE lines, with no difference between WT and OE fruits (Figure S9d). Furthermore, to investigate whether the increased firmness extended the shelf life of the fruit, we conducted postharvest behaviour assessments using WT, CR‐11 and CR‐13 fruits harvested at the RR stage. CR‐11 and CR‐13 fruits displayed increased resistance to postharvest deterioration, as indicated through measurement of water and weight loss (Figure 5d,e). Compared with the control, the content of protopectin was significantly increased, and the activity of pectinase was significantly decreased in CR lines (Figure 5f,g).
Figure 5.
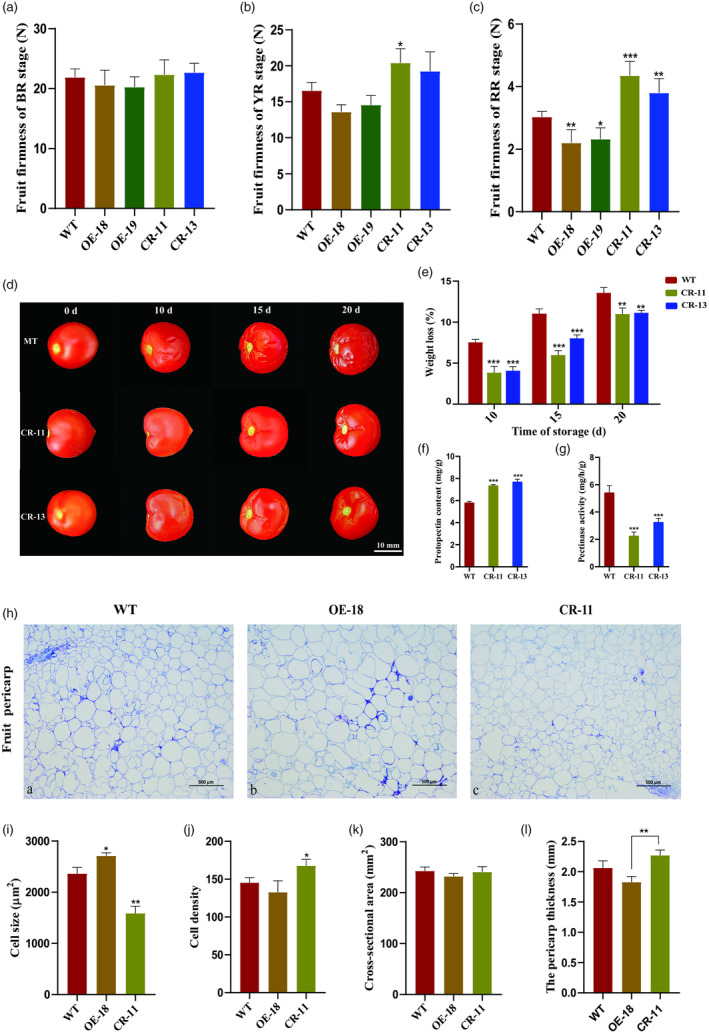
Fruit firmness, storage and pericarp microstructure of wild‐type (WT) and transgenic lines. (a‐c) Fruit firmness of WT and transgenic fruits at BR, YR and RR stages. (d) Fruits at the RR stage were harvested from WT, CR‐11 and CR‐13 plants and stored at room temperature for 20 days. Scale bar, 10 mm. (e) Physiological water loss (weight loss %) in WT, CR‐11 and CR‐13 fruits during different storage stages. The weight loss per fruit was calculated 10, 15 and 20 days after storage. (f) Protopectin content of WT, CR‐11 and CR‐13 fruits. (g) Pectinase activity of WT, CR‐11 and CR‐13 fruits. (h) Histological analysis of transverse sections of WT, OE‐18 and CR‐11 fruit pericarps at the red ripe stage. Scale bars = 500 μm. (i, j) Cell size and density of WT, OE‐18 and CR‐11 fruit pericarps. (k, l) Fruit cross‐section area and pericarp thickness of WT, OE‐18 and CR‐11 fruits. Values are presented as means ± SD (n = 10). Asterisks indicate the significant difference between WT and transgenic plants revealed by t‐test: *P < 0.05, **P < 0.01, ***P < 0.001.
To further understand the mechanism of SlEIN4 in regulating fruit firmness, we observed the pericarp tissue microstructure of WT and transgenic fruits. It can be found intuitively that the cell size of WT, CR‐11 and OE‐18 lines differed in the fruit pericarp tissue (Figure 5h). The average cell size of CR‐11 lines was significantly smaller than that of WT, while the OE‐18 line exhibited a significant increase compared with WT (Figure 5i). Moreover, the cell density of CR‐11 was significantly higher than that of the control. At the same time, OE‐18 showed a decrease with no significant difference from the control (Figure 5j). Meanwhile, the fruit cross‐sectional area analysis revealed no noticeable difference among WT, OE and CR lines (Figures 5k and S9a). Lastly, we measured the pericarp thickness of WT and transgenic lines. The result demonstrated that the pericarp thickness of OE‐18 decreased, while the pericarp thickness of CR‐11 increased compared with the control level. No clear difference between WT and transgenic lines was observed, but OE and CR lines showed a significant difference from each other (Figure 5l).
SNP (A → G) on SlEIN4 cause different genetic effects
To validate the SNP (A → G) effect on SlEIN4‐mediated fruit firmness, we overexpressed haplotype AA and GG SlEIN4 in MT tomato by transient transformation. Fruit transformation with empty pHellsgate8 was used as a control. Both were overexpressed significantly in the fruits (Figure 6b,c). Compared with the control, the fruits injected with pHellsgate8‐SlEIN4AA and pHellsgate8‐SlEIN4GG had a certain degree of delay in ripening (Figure 6a). The expression level of ERF genes was detected on the 14th day after transformation, and they decreased significantly in both pHellsgate8‐SlEIN4AA and pHellsgate8‐SlEIN4GG fruits compared with the control (Figure 6d,e). The result was consistent with the phenotype of overexpressed transgenic materials.
Figure 6.
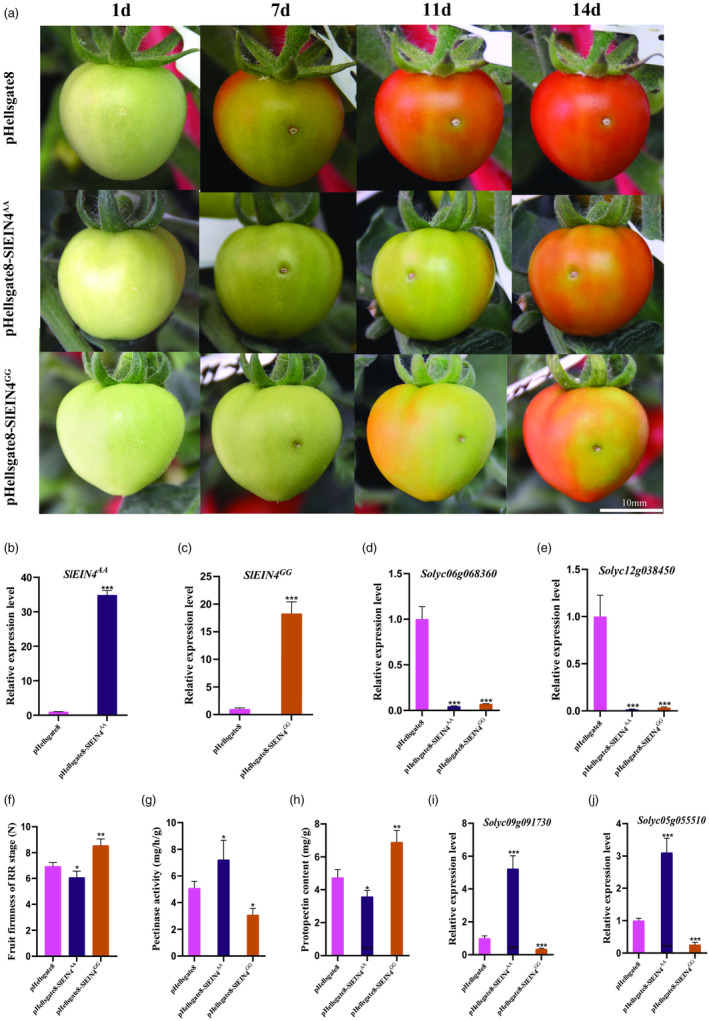
Phenotype, physiological data and related gene expression of pHellsgate8, phellsgate8‐SlEIN4AA and phellsgate8‐SlEIN4GG fruits. (a) Fruits phenotype from Day 1 to 14 after transient transformation. (b) The expression levels of SlEIN4 AA in phellsgate8‐SlEIN4AA fruits at Day 14 after transient transformation. (c) The expression levels of SlEIN4 GG in phellsgate8‐SlEIN4GG fruits at Day 14 after transient transformation. (d‐e) The relative expression levels of ERF genes in pHellsgate8, phellsgate8‐SlEIN4AA,\ and phellsgate8‐SlEIN4GG fruits at Day 14 after transient transformation. (f‐h) Fruit firmness, pectinase activity and protopectin content of pHellsgate8, phellsgate8‐SlEIN4AA and phellsgate8‐SlEIN4GG fruits at the RR stage. (i‐j) The relative expression level of pectinase genes in pHellsgate8, phellsgate8‐SlEIN4AA and phellsgate8‐SlEIN4GG fruits at the RR stage. Values are presented as means ± SD (n = 10). Asterisks indicate the significant difference between wild‐type and transgenic plants revealed by t‐test: *P < 0.05, **P < 0.01, ***P < 0.001.
In addition, the physiological characteristics of fruits were detected at the RR stage. Compared with the control, the firmness of pHellsgate8‐SlEIN4AA fruits decreased significantly, the pectinase activity was significantly increased, and the content of protopectin was significantly reduced (Figure 6f–h). Overexpression of SlEIN4 GG significantly increased fruit firmness, decreased pectinase activity and increased protopectin content (Figure 6f–h). The expression of pectinase genes in pHellsgate8‐EIN4AA fruits was increased, while these genes were decreased in pHellsgate8‐EIN4GG fruits compared with the control (Figure 6i,j). The above data suggested that the SNP (A → G) on SlEIN4 causes different genetic effects.
SlEIN4 gene editing impacts the expression of pectinase and ethylene regulatory pathway‐related genes
Transcriptome sequencing was conducted for both fruits at 39 dpa of WT and gene‐edited lines. The analysis revealed 1248 differentially expressed genes (DEGs), 585 were up‐regulated, and 663 were down‐regulated (Figure S10). The top 15 Gene Ontology (GO) pathways, distributed across biological processes, cellular components and molecular functions, are depicted in Figure 7a. Furthermore, the 1248 DEGs were enriched into five metabolic pathways, with the differential genes primarily enriched in pathways such as photosynthesis, photosynthesis‐antenna proteins, cutin, suberine and wax biosynthesis, fatty acid biosynthesis and phenylpropanoid biosynthesis (Figure 7b).
Figure 7.
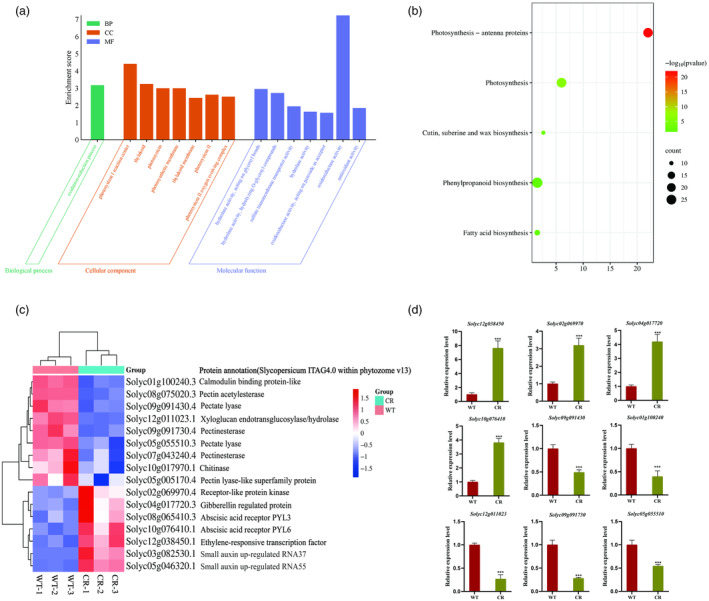
Transcriptome sequencing analysis of wild‐type (WT) and CR fruits at 39dpa. (a) Gene Ontology (GO) pathway annotation classification of differentially expressed genes (DEGs). The x‐coordinate represents the GO Term, and the y‐coordinate shows the GO Term enriched −log10 (P‐value), DEGs were selected with a value of | log2 (FoldChange) | >1 and P adj < 0.05, as indicated below. (b) Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment of differentially expressed genes. The horizontal coordinate represents the rich factor, and the vertical coordinate represents the pathway. The size of the dots in the figure corresponds to the number of differentially annotated genes in the corresponding pathway, and the depth of colours represents the significance level. (c) Clustering heatmap of DEGs related to pectinase and ethylene regulatory pathways. Genes are represented horizontally, with one sample for each column. Red indicates high expression, and blue indicates low expression. (d) The expression levels of pectinases genes, ethylene and GA regulatory pathway‐related genes in WT and CR lines. Values are presented as means ± SD (n = 3). Asterisks indicate the significant difference between wild‐type and transgenic plants revealed by t‐test: *P < 0.05, **P < 0.01, ***P < 0.001.
From the set of DEGs, we identified 16 candidate genes related to tomato firmness and ripening (Table S9). Among them, nine genes were associated with cell wall degradation, including two pectate lyase, two PE, one pectin acetylesterase, one PL‐like superfamily protein, one chitinase, one calmodulin binding protein‐like and one xyloglucan endotransglucosylase/hydrolase. The expression levels of these DEGs were predominantly down‐regulated in CR compared with WT (Figure 7c). Additionally, seven genes were related to ethylene regulatory pathways, including one ethylene‐responsive transcription factor, two abscisic acid receptors, one gibberellin‐regulated protein, two small auxin up‐regulated RNA, one receptor‐like protein kinase, all of which were found to participate in ethylene signal transduction or other hormonal pathways related to fruit development and maturation. The expression levels of these genes were up‐regulated mainly in CR compared with WT (Figure 7c). The expression levels of pectinase genes and ethylene, GA or ABA regulatory pathway‐related genes were further confirmed using qRT‐PCR. The results were consistent with the findings from the transcriptome sequencing (Figure 7d).
Discussion
Tomato fruit firmness plays a crucial role in its storage and transportation. Recent research has increasingly concentrated on understanding the regulation of fruit firmness, contributing to identifying several genes in this process. For instance, Yan et al. (2020) reported that BEL1‐LIKE HOMEODOMAIN4 (BLH4) participates in cell wall metabolism during tomato fruit ripening and is a crucial regulator of firmness. Additionally, SlBES1 has been found to promote tomato fruit softening by transcriptionally inhibiting pectin methylesterase ubiquitously 1 (PMEU1) (Liu et al., 2021). Fruit firmness is closely linked to fruit ripening, and previous studies have reported the impact of ethylene receptors on the ethylene regulation pathway, influencing fruit development and ripening (Anas et al., 2022; Chang et al., 1993; Kamiyoshihara et al., 2022; Shi et al., 2022). In our study, through GWAS, we successfully excavated a strong SNP peak related to fruit firmness. Specifically, we focused on an ethylene receptor, EIN4, which is predominantly expressed in fruits and contains a non‐synonymous SNP mutation, as indicated by linkage analysis. The SNP (A → G) of SlEIN4 was found in various materials, and intriguingly, haplotype AA was predominantly found in low‐firmness materials. In contrast, haplotype GG was mainly detected in high‐firmness materials, strongly suggesting SlEIN4 as a candidate gene associated with fruit firmness. Considering evolutionary perspectives, this variation is likely one of the factors contributing to the variation in tomato fruit firmness. To further validate the role of SlEIN4 (AA) in regulating tomato firmness, we employed MT tomato, which possesses the haplotype AA of the SlEIN4 gene, as the genetic background material. Gene editing of SlEIN4 accelerated tomato fruit ripening, whereas overexpression of this gene delayed fruit ripening compared with the control. Additionally, the ethylene release of CR was earlier than that of WT, while this of OE was later than that of WT. These observations provided the basis for earlier fruit ripening in gene‐edited lines and delayed ripening in overexpression lines. Based on previous research, CTR1, EIN2, EIN3 and ETR5 are known as essential regulatory genes in the ethylene pathway (Alonso et al., 1999; Chao et al., 1997; Kieber et al., 1993; Klee and Tieman, 2002). In this study, we also found that the expression levels of these genes were significantly altered in the fruit at the MG stage of the transgenic lines. The ethylene receptor is the receiver of the ethylene signal, located downstream of ethylene, so it cannot directly regulate ethylene production. It was speculated that the ethylene receptor regulates ethylene production by influencing the expression of downstream genes with feedback pathways.
According to a previous study, it was reported that ethylene‐responsive factor 4 (ERF4) binds to the promoter of ERF3, promoting ethylene synthesis and resulting in a decrease in fruit firmness (Hu et al., 2020). While a few genes in the ethylene regulation pathway have been reported to regulate firmness, whether ethylene receptors directly regulate fruit firmness remains largely unknown. In this research, we found that gene‐edited SlEIN4 enhanced the fruit firmness of tomatoes at the YR and RR stages, prolonging the shelf life of the fruits. Instead, overexpression of this gene reduced fruit firmness, indicating that SlEIN4 negatively regulated fruit firmness. To validate the SNP (A → G) effect on SlEIN4‐mediated firmness, we also overexpressed haplotype AA and GG SlEIN4 in MT tomato. The firmness of pHellsgate8‐EIN4AA fruits was significantly decreased while it was significantly increased in SlEIN4 GG overexpression fruit compared with control, suggesting the SNP (A → G) on SlEIN4 causes different genetic effects. In the 266 natural population, haplotype AA exists primarily in low‐firmness fruits, haplotype GG mainly exists in high‐firmness fruits, and heterozygous AG mainly in low‐firmness fruits. It could be inferred that haplotype AA is a dominant gene, and haplotype GG is a recessive gene. When the dominant gene was overexpressed, the firmness was reduced, and the firmness was increased when the gene was edited.
Fruit firmness is an intricate trait influenced by various characteristics of fruit cells, such as thickness, size and arrangement (Cao et al., 2012; Kendra and James, 2015; Lahaye et al., 2018). To further explore the mechanism of SlEIN4 in regulating fruit firmness, we investigated the fruit pericarp microstructure of WT and transgenic fruits. The pericarp cell size of CR lines was significantly decreased, while in the OE lines, it was noticeably increased compared with the control. It has been reported that larger pericarp cells are associated with lower fruit firmness (Cao et al., 2012; Vrebalov et al., 2002). Additionally, the cell density of CR lines was dramatically increased, while that of OE lines was decreased compared with the control. These data suggest that SlEIN4 likely regulates fruit firmness by simultaneously affecting pericarp cell size and cell density. Flesh firmness in tomatoes is closely linked to cell wall degradation. Enzymes such as PE, pectate lyase, PG, cellulase and chitinase participate in cell wall degradation, leading to the breakdown of fruit pectin, softening of fruit and a decrease in firmness (Su et al., 2022). RNA‐Seq analysis demonstrated that the transcription of most of the genes associated with cell wall degradation was down‐regulated in CR lines compared with the control, proving that cell wall degradation was hindered in CR lines. SlEIN4 AA positively regulated pectinase activity and pectinase gene expression while negatively regulating firmness and protopectin content. The genetic effects of SlEIN4 GG were the opposite.
Furthermore, several genes related to ethylene signal transduction and other hormonal pathways, such as ethylene‐responsive transcription factor, receptor‐like protein kinase, abscisic acid receptor and gibberellin‐regulated protein mainly were up‐regulated in the CR lines compared with WT, resulting in an advancement in fruit maturity in CR lines. The ethylene regulatory network is complex, and other hormones related to development and maturation, such as GA and ABA, also play a role and can cooperate with ethylene regulation (Li et al., 2022; Wu et al., 2023). Our results suggested that SlEIN4 does influence fruit firmness to some extent. However, the exact mechanism of how SlEIN4 regulates fruit firmness needs further investigation.
In conclusion, this study represents the first report demonstrating that an ethylene receptor gene can regulate fruit firmness by affecting cell wall degradation and fruit pericarp microstructure. Our findings offered a valuable genetic understanding of the natural variability of tomato fruit firmness and maturity, which can further facilitate the effective use of these traits in tomato improvement.
Experimental procedures
Plant materials and sequencing
GWAS was conducted using a diverse collection of 266 tomato resources, comprising 149 large tomatoes (S. l. lycopersicum, BIG), 103 cherry tomatoes (var cerasiforme, CER) and 14 currant tomatoes (Solanum pimpinellifolium, PIM), which were carefully selected from a larger pool of 360 natural population resources reported in a previous study and sourced from various global regions (Lin et al., 2014). The experimental population material was cultivated at two distinct locations in Wuhan, China, in 2013: the HZ and ZD locations. During GWAS, we harvested at least six fruits from at least six plants per row at the RR stage to test for fruit firmness.
Genome read mapping and variant calling
We obtained the reads dataset of 266 tomato accessions, which had been previously reported, from the Sol Genomics Network (https://solgenomics.net/). Using the burrows‐wheeler aligner (BWA) (v0.7.12) with default parameters, the reads were cleaned and aligned to the ‘Heinz 1706’ tomato genome version SL2.4 (https://solgenomics.net/). Subsequently, duplicated read pairs were identified using Picard (v2.0.1) with default parameters. For variant calling, we utilized the HaplotypeCaller function of the genome analysis toolkit (GATK) (McKenna et al., 2010) (version 20 171 018) to generate genome variant call format (GVCF) files for each accession. Then, we performed population variant calling with the GenotypeGVCFs function of GATK, employing the default parameters. Finally, we applied hard filtering to the raw variant set using GATK, with parameters ‘QD<2.0 || FS> 60.0|| MQ< 40.0 || MQRankSum<−12.5 || ReadPosRankSum<−8.0’ applied to SNPs, and ‘QD < 2.0 || FS > 200.0 || ReadPosRankSum<−20.0’ applied to small indels.
Phylogenetic and population analyses
For population genomic analyses, we retained bi‐allelic SNPs with a missing data rate of <15% and a minor allele frequency (MAF) > 0.05. PCA was conducted using Plink (Purcell et al., 2007) (v1.9) with the complete set of SNPs (5 485 483 SNPs). We employed STRUCTURE (Falush et al., 2003) (v2.3.4) to assess population structure. The optimal number of clusters (K) was determined, showing the most probable quantity of population clusters. Using 8000 randomly selected SNPs at fourfold degenerated sites, STRUCTURE analyses were run 20 times for each K value ranging from 2 to 20. The optimal K was determined as 3 by following the ΔK method (Evanno et al., 2005) implemented in STRUCTURE. The PCA plots and population structure graph (for each K = 2–4) were drawn with R packages. The Balding–Nichols kinship matrix (BN matrix) was constructed in emmax‐kin and the proportion of SNPs was randomly selected with the default parameter (emmax‐kin ‐v ‐h ‐d 10). The heatmap of the kinship matrix among 266 tomato access was drawn by the pheatmap package (https://cran.r‐project.org/web/packages/pheatmap/) in R.
Linkage disequilibrium analysis
Haploview 4.2 software was used to analyse LD (Barrett et al., 2005). Specifically, we evaluated the average linkage decay along each 0.5‐Mb region of the entire genome, parameters set as: ‐maxdistance 500 ‐minMAF 0.05 ‐hwcutoff 0. To identify the physical locations of the SNPs, we referred to the tomato genomic sequence version SL2.40 (http://solgenomics.net/).
Genome‐wide association study
A total of 5 485 483 SNPs consist in the GWAS analysis, with a minor allele frequency of 0.01 or higher and a lost data rate of 15% or less across the total population. The GWAS was conducted using a mixed linear model (MLM) fitted in the GEMMA algorithm (Zhou and Stephens, 2012). The MLM considers kinship (BN matrix calculated by emmax‐kin) and population structure (Q matrix) to control for potential confounding factors. The P‐value was counted for each SNP, and statistical significance was determined using a uniform threshold of ≤1.8 × 10−7 (P = 1/n; n = total number of markers used) as the cut‐off for identifying significant associations with the firmness traits (Wen et al., 2014). An R package qqman (https://cran.r‐project.org/web/packages/qqman/) was used to create (quantile‐quantile) Q‐Q and Manhattan plots from GWAS results.
RNA isolation and gene expression analysis
Tomato plant RNA was extracted and purified following the Hua Yue Yang kit's instructions. Genomic DNA was removed using Monad's reverse transcription reagent, and the remaining RNA was reverse transcribed to generate cDNA. For qRT‐PCR, we used Primer Premier 5.0 design primers. The experiments used 2 × HQ SYBR qPCR Mix (ZOMANBIO, China) and the CFX96 real‐time PCR system (Bio‐Rad, USA). The reference gene used was Solyc11g005330. Each qRT‐PCR experiment was performed in triplicate as three biological replicates. The relative gene expression was determined using the 2−▵▵Ct approach. The primer sequences used for qRT‐PCR are found in Table S6.
Subcellular localization
PCR was utilized to amplify the coding sequence (CDS) fragment of SlEIN4, ensuring the absence of a stop codon. Subsequently, the ClonExpress II One Step Cloning Kit was employed to insert the amplified fragment into the pCAMBIA1302 vector (Vazyme, China). The pCAMBIA1302‐SlEIN4‐GFP with cell membrane marker Cd‐1007‐mCherry (Yang et al., 2018) and the control vector (pCAMBIA1302‐GFP) with Cd‐1007‐mCherry were subsequently infiltrated into approximately 4‐week‐old Nicotiana benthamiana leaves by introduced into Agrobacterium tumefaciens GV3101 (Batistic et al., 2010). After 48 h of infiltration, protoplasts were prepared from infiltrated tobacco leaves using a protoplast extraction kit (Coolaber, China). Green fluorescent protein (GFP) and mCherry fluorescence were visualized using a laser confocal microscope (Nikon, Japan).
Amino acid sequence alignment and phylogenetic analysis
The full‐length amino acid sequences of EIN4 and SlETR1‐SlETR7 homologous genes from tomatoes were obtained from the NCBI public database (https://www.ncbi.nlm.nih.gov/). DNAMAN software was used to compare the amino acid sequences obtained. Molecular evolutionary genetics analysis (MEGA) 7 software was used to construct the evolutionary tree, and the result was obtained by further calculation.
Plant transformation
The full‐length SlEIN4 open reading frame (ORF) was amplified from tomato MT cDNA and integrated into the pHELLSGATE8 vector through homologous recombination (ClonExpress II One Step Cloning Kit, Vazyme). Two specific targets (sgRNA1 and sgRNA2) were manually chosen for the gene editing process for SIEIN4. The CP098 recombinant vector was designed to incorporate two specific sgRNAs targeting the SlEIN4 CDS, resulting in a defined deletion of the Cas9 endonuclease gene (Ye et al., 2017). All primers utilized for vector construction are listed in Table S4. The plasmid was transferred into the tomato genome via cotyledon explants by introducing it into Agrobacterium tumefaciens strain GV3101. Micro Tom was the chosen plant for the transformation process. Positive T0 plants were certified by PCR using CRISPR/Cas9 detection primers (Table S5). CRISPR/Cas9‐induced mutations were further characterized using PCR sequencing employing CR‐SlEIN4‐specific primers (Table S5). Overexpression plants were verified through PCR using CaMV35S promoter forward primers and OE‐SlENI4‐specific reverse primers (Table S5). Both OE and CR transgenic lines used in this research were homozygous lines. The SIEIN4 promotor sequence from MT was inserted into a pMV2 vector harbouring the β‐glucosidase (GUS) gene, resulting in the ProSlEIN4::GUS vector. This construct was then transformed into MT.
Tomato transient transformation
The full‐length Haplotype GG of SlEIN4 ORF was amplified out of tomato M82 (TS‐3) cDNA and integrated into the pHELLSGATE8 vector (Figure S11). The recombinant vector was named pHellsgate8‐SlEIN4GG. pHellsgate8‐SlEIN4AA was the same vector transformed into OE. Recombinant vectors and pHellsgate8 empty vector were transformed into Agrobacterium tumefaciens strain GV3101. pHellsgate8 empty vector was used as a control. MT fruits at the MG stage (34dpa) were used in the transient expression experiments following the method of Zhang and Duan (Duan et al., 2022; Zhang et al., 2015). A single fruit was considered a biological replicate, and at least six replicates were taken.
Determination of ethylene production, fruit firmness and other fruit quality traits
To assess ethylene production and ACC content in fruit, we collected tomato fruits from WT, OE and CR lines at various ripening stages, including 31 dpa, 34 dpa, 37 dpa, 39 dpa and 41 dpa. The ethylene measurement method followed the protocol described by Gao (Gao et al., 2018). The method for determination of ACC content in fruit followed the protocol described by Kazmierczak and Petritis (Kazmierczak and Kazmierczak, 2007; Petritis et al., 2003). For firmness measurement, we used a texture analyser TA.XTPlus (Godalming, UK) to determine the fruit firmness in Newtons (N).
The ripening time of the fruits was determined by counting the number of days from flowering to the RR stage. The fruit shape index was defined as the longitudinal diameter divided by the transverse diameter. Pericarp thickness was measured using digital vernier callipers (Guang Lu, China) from the central cavity to the outer peel. Fruit weight was assessed using an electronic balance (YOKE, China). For chemical analysis, the titratable acidity of the fruits was estimated following the method supplied by Vijayakumar et al. (2021), and soluble sugar content was measured as per the procedure outlined by Hu et al. (2015). For the microscopic examination of pericarp tissue, paraffin sections were prepared using the method described by Li et al. (2019). The protopectin content was determined using the protopectin assay kit (Michy Biomedical Technology, China), and the fruit pectinase activity was detected using a pectinase assay kit (Michy Biomedical Technology, China). A single fruit was considered a biological replicate, and at least 10 replicates were taken.
RNA‐Seq analysis
The samples of MT and SlEIN4‐CR fruits at 39 days post‐flowering were taken for transcriptomic analysis, respectively; each line has three replicates. The purity, concentration and integrity of total RNA were checked, and the libraries were produced, sequenced and analysed by Shanghai Personal Biotechnology Co. Ltd. Heatmap of DEGs using TBtools software. The sequencing data were of good quality and satisfied the subsequent analysis conditions (Tables S7 and S8). The conditions for screening differentially expressed genes were as follows: expression difference multiple |log2FoldChange| > 1, with adjusted P‐value (P adj) < 0.05 (P‐value was calculated by negative binomial distribution method). topGO was used for GO enrichment analysis, and the P‐value was calculated by the hypergeometric distribution method (the standard for significant enrichment was P adj < 0.05) to determine the GO term of significant enrichment of differential genes to assess the main biological functions of differential genes. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed using clusterProfiler (3.4.4) software, focusing on significant enrichment pathways with P adj < 0.05. Finally, an online mapping website (https://www.bioinformatics.com.cn/) was used for drawing.
Statistical analysis
All experiments proceeded independently and were repeated in triplicate to ensure reproducibility. Data analysis was executed using SPSS (SPSS Corporation, Chicago, USA) with analysis of variance procedures. Significant differences among the means were assessed using Student's t‐test at the P < 0.05, P < 0.01 and P < 0.001 levels. The reported values represent the mean ± standard deviation.
Author contributions
S.Z., W.W. and Z.Y. designed the research. S.Z., S.W., B.F. and X.M. performed experiments. Z.J. and Y.L. provided the background material. S.Z., J.Z. and P.W. analysed the GWAS data. S.Z., S.W. and Y.G. analysed the experimental data. S.Z. and W.W. wrote and revised the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Figure S1 Population structure analysis of 266 tomato natural population GWAS panels based on 5 485 483 SNPs. (a) A phylogenetic matrix diagram displays the relationships between populations. The X‐axis and the right Y‐axis show individual plants, while the left Y‐axis represents a tree plot illustrating phylogenetic relationships derived from SNP data. (b) Population structure of tomato germplasm with varying cluster numbers (K = 2, 3 and 4). The x‐axis denotes different groups and their respective positions. (c–d) PCA plot of the tomato accessions, with the percentage of variance explained by each PC indicated in parenthesis. PC1, the first principal component; PC2, the second principal component; PC3, the third principal component.
Figure S2 The conserved domain in SlEIN4. One histidine kinase domain is predicted using the InterPro database.
Figure S3 SlEIN4 gene sequence in wild‐type tomato.
Figure S4 Phylogenetic tree analysis of SlEIN4 and SlETR1‐SlETR7 proteins in tomato. The scale bar presents the substitution per amino acid based on the bootstrap method (B = 1000 replications).
Figure S5 Amino acid sequence alignment of SlEIN4 and SlETR1‐SlETR7 in tomato. Black shade indicates amino acid residues 100% conserved, pink shade indicates amino acids that are >80% conserved, and blue shade indicates amino acids that are >50% conserved.
Figure S6 PCR genotyping of two T0 generation CR‐SlEIN4 mutants, PCR amplicon of Cas9, is a positive control.
Figure S7 Sequencing analysis of CR‐SlEIN4 alleles sequences from T2 homozygous lines of CR‐11 and CR‐13.
Figure S8 Fruit ripening phenotypes of WT, OE‐18 and CR‐11 plants. Scale bar, 2 cm.
Figure S9 Transverse section and quality of WT and transgenic fruits. (a) Transverse section of WT, OE‐18 and CR‐11 fruits. Scale bar, 10 mm. (b‐d) Fruit weight, sugar‐acid ratio and total soluble solids of WT and transgenic fruits at red ripe stage. Values are presented as means ± SD (n = 10). Asterisks indicate the significant difference between wild‐type and transgenic plants revealed by t‐test: *P < 0.05, **P < 0.01, ***P < 0.001.
Figure S10 Volcano map and clustering heatmap of differentially expressed genes (DEGs) between WT and CR lines. (a) Volcano map. The x‐coordinate represents the log2 (FoldChange), and the y‐coordinate represents the negative log value of 10 at the significance level. The two dotted lines in the figure represent the threshold values of the difference multiples, the dotted line is the threshold of significance level. The colour indicates whether the gene is up‐regulated, down‐regulated or insignificantly differentially expressed (DEGs were selected with a value of | log2 (FoldChange) | >1 and P adj < 0.05, as indicated below). (b) Clustering heatmap. Genes are represented horizontally, with one sample for each column. Red indicates high expression, and blue indicates low expression.
Figure S11 Sequencing of SlEIN4 haplotype in tomato varieties MT and M82.
Table S1 The geographic distribution of SNP (ch04_55126611) in 266 tomato accessions.
Table S2 List of 50 SNPs significantly associated with tomato fruit firmness.
Table S3 Genes included in the location of 55 127 814–55 157 228.
Table S4 Primers used for vector constructions.
Table S5 Primers used for positive and gene sequencing detection of transgenic plants.
Table S6 Primers used for qRT‐PCR.
Table S7 Reads quality test.
Table S8 Clean Reads comparison of reference genomes.
Table S9 Expression analysis of DEGs related to pectinase, ethylene or other hormone regulatory pathways.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (31902024), the Scientific and Technological Project of Henan Province (212102110413, 202102110402) and the Research Fund for the Doctoral Program of Henan Agricultural University (30500495).
Contributor Information
Zhibiao Ye, Email: zbye@mail.hzau.edu.cn.
Wei Wang, Email: wangwei86@henau.edu.cn.
Data availability
The data used to support the findings of this study are included in the article and the supporting information file.
References
- Adams‐Phillips, L. , Barry, C. and Giovannoni, J. (2004) Signal transduction systems regulating fruit ripening. Trends Plant Sci. 9, 331–338. [DOI] [PubMed] [Google Scholar]
- Alexander, L. and Grierson, D. (2002) Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J. Exp. Bot. 53, 2039–2055. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M. , Hirayama, T. , Roman, G. , Nourizadeh, S. and Ecker, J.R. (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science, 284, 2148–2152. [DOI] [PubMed] [Google Scholar]
- Anas, A. , Wiguna, G. , Damayanti, F. , Mubarok, S. , Setyorini, D. and Ezura, H. (2022) Effect of ethylene sletr1‐2 receptor allele on flowering, fruit phenotype, yield, and shelf‐life of four F1 generations of tropical tomatoes (Solanum lycopersicum L.). Horticulturae, 8, 1098. [Google Scholar]
- Atkinson, R.G. , Sutherland, P.W. , Johnston, S.L. , Gunaseelan, K. , Hallett, I.C. , Mitra, D. , Brummell, D.A. et al. (2012) Down‐regulation of POLYGALACTURONASE1 alters firmness, tensile strength and water loss in apple (Malus x domestica) fruit. BMC Plant Biol. 12, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett, J.C. , Fry, B. , Maller, J. and Daly, M.J. (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics, 21, 263–265. [DOI] [PubMed] [Google Scholar]
- Batistic, O. , Waadt, R. , Steinhorst, L. , Held, K. and Kudla, J. (2010) CBL‐mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J. 61, 211–222. [DOI] [PubMed] [Google Scholar]
- Brummell, D.A. and Harpster, M.H. (2001) Cell wall metabolism in fruit softening and quality and its manipulation in transgenic plants. Plant Mol. Biol. 47, 311–340. [PubMed] [Google Scholar]
- Cao, Y. , Tang, X.F. , Giovannoni, J. , Xiao, F.M. and Liu, Y.S. (2012) Functional characterization of a tomato COBRA‐like gene functioning in fruit development and ripening. BMC Plant Biol. 12, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C. , Kwok, S.F. , Bleecker, A.B. and Meyerowitz, E.M. (1993) Arabidopsis ethylene‐response gene ETR1: similarity of product to two‐component regulators. Science, 262, 539–544. [DOI] [PubMed] [Google Scholar]
- Chao, Q. , Rothenberg, M. , Solano, R. , Roman, G. , Terzaghi, W. and Ecker, J.R. (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein THYLENE‐INSENSITIVE3 and related proteins. Cell, 89, 1133–1144. [DOI] [PubMed] [Google Scholar]
- Chapman, N.H. , Bonnet, J. , Grivet, L. , Lynn, J. , Graham, N. , Smith, R. , Sun, G. et al. (2012) High‐resolution mapping of a fruit firmness‐related quantitative trait locus in tomato reveals epistatic interactions associated with a complex combinatorial locus. Plant Physiol. 159, 1644–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Hu, G. , Rodriguez, C. , Liu, M. and Chervin, C. (2020) Roles of SlETR7, a newly discovered ethylene receptor, in tomato plant and fruit development. Hortic. Res. 7, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, X.Y. , Cai, J. , Zhao, Y.P. , Gao, G. , Li, M. and Qi, H.Y. (2022) Transcriptome and metabolomics analysis revealed that CmWRKY49 regulating CmPSY1 promotes β‐carotene accumulation in orange fleshed oriental melon. Hortic. Plant J. 8, 650–666. [Google Scholar]
- Evanno, G. , Regnaut, S. and Goudet, J. (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 14, 2611–2620. [DOI] [PubMed] [Google Scholar]
- Falush, D. , Stephens, M. and Pritchard, J.K. (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics, 164, 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frary, A. , Fulton, T.M. , Zamir, D. and Tanksley, S.D. (2004) Advanced backcross QTL analysis of a Lycopersicon esculentum × L. pennellii cross and identification of possible orthologs in the Solanaceae. Theor. Appl. Genet. 108, 485–496. [DOI] [PubMed] [Google Scholar]
- Fulton, T.M. , Beck‐Bunn, T. , Emmatty, D. , Eshed, Y. , Lopez, J. , Petiard, V. , Uhlig, J. et al. (1997) QTL analysis of an advanced backcross of Lycopersicon peruvianum to the cultivated tomato and comparisons with QTLs found in other wild species. Theor. Appl. Genet. 95, 881–894. [Google Scholar]
- Gao, Y. , Wei, W. , Zhao, X. , Tan, X.L. , Fan, Z.Q. , Zhang, Y.P. , Jing, Y. et al. (2018) A NAC transcription factor, NOR‐like1, is a new positive regulator of tomato fruit ripening. Hortic. Res. 5, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapper, N.E. , McQuinn, R.P. and Giovannoni, J.J. (2013) Molecular and genetic regulation of fruit ripening. Plant Mol. Biol. 82, 575–591. [DOI] [PubMed] [Google Scholar]
- Hu, Y. , Wu, Q. , Sprague, S.A. , Park, J. , Oh, M. , Rajashekar, C.B. , Koiwa, H. et al. (2015) Tomato expressing Arabidopsis glutaredoxin gene AtGRXS17 confers tolerance to chilling stress via modulating cold responsive components. Hortic. Res. 2, 15051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y.N. , Han, Z.Y. , Sun, Y.Q. , Wang, S. , Wang, T. , Wang, Y. , Xu, K.N. et al. (2020) ERF4 affects fruit firmness through TPL4 by reducing ethylene production. Plant J. 103, 937–950. [DOI] [PubMed] [Google Scholar]
- Hua, J. and Meyerowitz, E.M. (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana . Cell, 94, 261–271. [DOI] [PubMed] [Google Scholar]
- Huang, W. , Hu, N. , Xiao, Z.N. , Qiu, Y.P. , Yang, Y. , Yang, J. , Mao, X. et al. (2022) A molecular framework of ethylene‐mediated fruit growth and ripening processes in tomato. Plant Cell, 34, 3280–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiyoshihara, Y. , Achiha, Y. , Ishikawa, S. , Mizuno, S. , Mori, H. , Tateishi, A. , Huber, D.J. et al. (2022) Heteromeric interactions of ripening‐related ethylene receptors in tomato fruit. J. Exp. Bot. 73, 6773–6783. [DOI] [PubMed] [Google Scholar]
- Kazmierczak, A. and Kazmierczak, J.M. (2007) The level of endogenous 1‐aminocyclopropane‐1 ‐carboxylic acid in gameto‐phytes of Anemia phyllitidis is increased during GA3‐induced antheridia formation. Acta Physiol. Plant. 29, 211–216. [Google Scholar]
- Kendra, M.B. and James, W.O. (2015) Cell wall composition of the skin and flesh tissue of crisp and standard texture southern highbush blueberry genotypes. J. Berry Res. 5, 9–15. [Google Scholar]
- Kevany, B.M. , Tieman, D.M. , Taylor, M.G. , Dal Cin, V. and Klee, H.J. (2007) Ethylene receptor degradation controls the timing of ripening in tomato fruit. Plant J. 51, 458–467. [DOI] [PubMed] [Google Scholar]
- Kevany, B.M. , Taylor, M.G. and Klee, H.J. (2008) Fruit‐specific suppression of the ethylene receptor LeETR4 results in early‐ripening tomato fruit. Plant Biotechnol. J. 6, 295–300. [DOI] [PubMed] [Google Scholar]
- Kieber, J.J. , Rothenberg, M. , Roman, G. , Feldmann, K.A. and Ecker, J.R. (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell, 72, 427–441. [DOI] [PubMed] [Google Scholar]
- Klee, H. and Tieman, D. (2002) The tomato ethylene receptor gene family: form and function. Physiol. Plant. 115, 336–341. [DOI] [PubMed] [Google Scholar]
- Lahaye, M. , Bouin, C. , Barbacci, A. , Le Gall, S. and Foucat, L. (2018) Water and cell wall contributions to apple mechanical properties. Food Chem. 268, 386–394. [DOI] [PubMed] [Google Scholar]
- Li, L. , Zhao, W. , Feng, X. , Chen, L.L. , Zhang, L.D. and Zhao, L.X. (2019) Changes in fruit firmness, cell wall composition, and transcriptional profile in the yellow fruit tomato 1 (yft1) mutant. J. Agric. Food Chem. 67, 463–472. [DOI] [PubMed] [Google Scholar]
- Li, R. , Sun, S. , Wang, H.J. , Wang, K.T. , Yu, H. , Zhou, Z. , Xin, P.Y. et al. (2020) FIS1 encodes a GA2‐oxidase that regulates fruit firmness in tomato. Nat. Commun. 11, 5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B.J. , Grierson, D. , Shi, Y.N. and Chen, K.S. (2022) Roles of abscisic acid in regulating ripening and quality of strawberry, a model non‐climacteric fruit. Hortic. Res. 9, uhac089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, T. , Zhu, G.T. , Zhang, J.H. , Xu, X.Y. , Yu, Q.H. , Zheng, Z. et al. (2014) Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 46, 1220–1226. [DOI] [PubMed] [Google Scholar]
- Liu, Q. and Wen, C.K. (2012) Arabidopsis ETR1 and ERS1 differentially repress the ethylene response in combination with other ethylene receptor genes. Plant Physiol. 158, 1193–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H.R. , Liu, L.H. , Liang, D.Y. , Zhang, M. , Jia, C.G. , Qi, M.F. , Liu, Y.Y. et al. (2021) SlBES1 promotes tomato fruit softening through transcriptional inhibition of PMEU1. iScience, 24, 102926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, N. , Feng, H. , Meng, X. , Li, D. , Yang, D. , Wu, C. and Meng, Q. (2014) Overexpression of tomato SlNAC1 transcription factor alters fruit pigmentation and softening. BMC Plant Biol. 14, 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna, A. , Hanna, M. , Banks, E. , Sivachenko, A. , Cibulskis, K. , Kernytsky, A. , Garimella, K. et al. (2010) The genome analysis toolkit: a MapReduce framework for analyzing next‐generation DNA sequencing data. Genome Res. 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheli, F. (2001) Pectin methylesterases: Cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 6, 414–419. [DOI] [PubMed] [Google Scholar]
- Monsalve, L. , Bernales, M. , Ayala‐Raso, A. , Alvarez, F. , Valdenegro, M. , Alvaro, J.E. , Figueroa, C.R. et al. (2022) Relationship between Endogenous Ethylene Production and Firmness during the Ripening and Cold Storage of Raspberry (Rubus idaeus ‘Heritage’) Fruit. Horticulturae, 8, 262. [Google Scholar]
- Mubarok, S. , Hoshikawa, K. , Okabe, Y. , Yano, R. , Tri, M.D. , Ariizumi, T. and Ezura, H. (2019) Evidence of the functional role of the ethylene receptor genes SlETR4 and SlETR5 in ethylene signal transduction in tomato. Mol. Genet. Genomics, 294, 301–313. [DOI] [PubMed] [Google Scholar]
- Nordborg, M. and Weigel, D. (2008) Next‐generation genetics in plants. Nature, 456, 720–723. [DOI] [PubMed] [Google Scholar]
- Oetiker, J.H. and Yang, S.F. (1995) The role of ethylene in fruit ripening. Acta Hortic. 398, 167–178. [Google Scholar]
- Petritis, K. , Koukaki, G. , Koussissi, E. , Elfakir, C. , Dreux, M. and Dourtoglou, V. (2003) The simultaneous determination of 1‐aminocyclopropane‐1‐carboxylic acid and cyclopropane‐1,1‐dicarboxylic acid in Lycopersicum esculentum by high performance liquid chromatography‐electrospray tandem mass spectrometry. Phytochem. Anal. 14, 347–351. [DOI] [PubMed] [Google Scholar]
- Purcell, S. , Neale, B. , Todd‐Brown, K. , Thomas, L. , Ferreira, M.A.R. , Bender, D. , Maller, J. et al. (2007) PLINK: A tool set for whole‐genome association and population‐based linkage analyses. Am. J. Hum. Genet. 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y. , Vrebalov, J. , Zheng, H. , Xu, Y.M. , Yin, X.R. , Liu, W.L. , Liu, Z.M. et al. (2021) A tomato LATERAL ORGAN BOUNDARIES transcription factor, SlLOB1, predominantly regulates cell wall and softening components of ripening. Proc. Natl. Acad. Sci. U. S. A. 118, e2102486118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J.Y. , Wang, Q. , Zuo, J.H. , Zheng, S.F. , Gao, L.P. , Liu, J. and Wang, Q. (2022) Comparative proteomic analysis of wild‐type and a SlETR‐3 (Nr) mutant reveals an ethylene‐induced physiological regulatory network in fresh‐cut tomatoes. Food Res. Int. 161, 111491. [DOI] [PubMed] [Google Scholar]
- Stepanova, A.N. and Ecker, J.R. (2000) Ethylene signaling: from mutants to molecules. Curr. Opin. Plant Biol. 3, 353–360. [DOI] [PubMed] [Google Scholar]
- Su, Q. , Li, X. , Wang, L. , Wang, B. , Feng, Y. , Yang, H. and Zhao, Z. (2022) Variation in cell wall metabolism and flesh firmness of four apple cultivars during fruit development. Foods, 11, 3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thole, V. , Vain, P. , Yang, R. , Silva, J.A.B.D. and Martin, C. (2020) Analysis of tomato postharvest properties: fruit color, shelf life, and fungal susceptibility. Curr. Protoc. Plant Biol. 5, e20108. [DOI] [PubMed] [Google Scholar]
- Tieman, D.M. , Ciardi, J.A. , Taylor, M.G. and Klee, H.J. (2001) Members of the tomato LeEIL (EIN3‐like) gene family are functionally redundant and regulate ethylene responses throughout plant development. Plant J. 26, 47–58. [DOI] [PubMed] [Google Scholar]
- Usenik, V. , Kastelec, D. , Veberič, R. and Štampar, F. (2008) Quality changes during ripening of plums (Prunus domestica L.). Food Chem. 111, 830–836. [Google Scholar]
- Vijayakumar, A. , Shaji, S. , Beena, R. , Sarada, S. , Rani, T.S. , Stephen, R. , Manju, R.V. et al. (2021) High temperature induced changes in quality and yield parameters of tomato (Solanum lycopersicum L.) and similarity coefficients among genotypes using SSR markers. Heliyon, 7, e05988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent, H. , Wiersema, J. , Kell, S. , Fielder, H. , Dobbie, S. , Castaneda‐Alvarez, N.P. , Guarino, L. et al. (2013) A prioritized crop wild relative inventory to help underpin global food security. Biol. Conserv. 167, 265–275. [Google Scholar]
- Vrebalov, J. , Ruezinsky, D. , Padmanabhan, V. , White, R. , Medrano, D. , Drake, R. , Schuch, W. et al. (2002) A MADs‐Box gene necessary for fruit ripening at the tomato ripening‐inhibitor (Rin) locus. Science, 296, 343–346. [DOI] [PubMed] [Google Scholar]
- Wakabayashi, K. , Hoson, T. and Huber, D.J. (2003) Methyl‐de‐esterification as a major factor regulating the extent of pectin depolymerization during fruit ripening: a comparison of the action of avocado (Persea americana) and tomato (Lycopersicon esculentum) polygalacturonases. J. Plant Physiol. 160, 667–673. [DOI] [PubMed] [Google Scholar]
- Wang, K.L.C. , Li, H. and Ecker, J.R. (2002) Ethylene biosynthesis and signaling networks. Plant Cell, 14, S131–S151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D. , Samsulrizal, N.H. , Yan, C. , Allcock, N.S. , Craigon, J. , Blanco‐Ulate, B. , Ortega‐Salazar, I. et al. (2019) Characterization of CRISPR mutants targeting genes modulating pectin degradation in ripening tomato. Plant Physiol. 179, 544–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, W. , Li, D. , Li, X. , Gao, Y. , Li, W. , Li, H. , Liu, J. et al. (2014) Metabolome‐based genome‐wide association study of maize kernel leads to novel biochemical insights. Nat. Commun. 5, 3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, J.Q. , Lanahan, M.B. , Yen, H.C. , Giovannoni, J.J. and Klee, H.J. (1995) An ethylene‐inducible component of signal transduction encoded by Never‐ripe. Science, 270, 1807–1809. [DOI] [PubMed] [Google Scholar]
- Wu, M. , Liu, K.D. , Li, H.H. , Li, Y. , Zhu, Y.Q. , Su, D. , Zhang, Y.X. et al. (2023) Gibberellins involved in fruit ripening and softening by mediating multiple hormonal signals in t\omato. Hortic. Res. 11, uhad275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, F. , Gao, Y.S. , Pang, X.Q. , Xu, X. , Zhu, N. , Chan, H.E. , Hu, G.J. et al. (2020) BEL1‐LIKE HOMEODOMAIN4 regulates chlorophyll accumulation, chloroplast development, and cell wall metabolism in tomato fruit. J. Exp. Bot. 71, 5549–5561. [DOI] [PubMed] [Google Scholar]
- Yang, S.F. and Hoffman, N.E. (1984) Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Biol. 35, 155–189. [Google Scholar]
- Yang, S. , Cai, Y.L. , Liu, X.W. , Dong, M.M. , Zhang, Y.Q. , Chen, S.Y. , Zhang, W.B. et al. (2018) A CsMYB6‐CsTRY module regulates fruit trichome initiation in cucumber. J. Exp. Bot. 69, 1887–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, J. , Wang, X. , Hu, T. , Zhang, F. , Wang, B. , Li, C. , Yang, T.X. et al. (2017) An InDel in the promoter of Al‐ACTIVATED MALATE TRANSPORTER9 selected during tomato domestication determines fruit malate contents and aluminum tolerance. Plant Cell, 29, 2249–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, N. , Jiang, J. , Yl, Y. and Wang, Z.H. (2015) Functional characterization of an invertase inhibitor gene involved in sucrose metabolism in tomato fruit. J. Zhejiang Univ. Sci. B. 16, 845–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X. and Stephens, M. (2012) Genome‐wide efficient mixed‐model analysis for association studies. Nat. Genet. 44, 821–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Population structure analysis of 266 tomato natural population GWAS panels based on 5 485 483 SNPs. (a) A phylogenetic matrix diagram displays the relationships between populations. The X‐axis and the right Y‐axis show individual plants, while the left Y‐axis represents a tree plot illustrating phylogenetic relationships derived from SNP data. (b) Population structure of tomato germplasm with varying cluster numbers (K = 2, 3 and 4). The x‐axis denotes different groups and their respective positions. (c–d) PCA plot of the tomato accessions, with the percentage of variance explained by each PC indicated in parenthesis. PC1, the first principal component; PC2, the second principal component; PC3, the third principal component.
Figure S2 The conserved domain in SlEIN4. One histidine kinase domain is predicted using the InterPro database.
Figure S3 SlEIN4 gene sequence in wild‐type tomato.
Figure S4 Phylogenetic tree analysis of SlEIN4 and SlETR1‐SlETR7 proteins in tomato. The scale bar presents the substitution per amino acid based on the bootstrap method (B = 1000 replications).
Figure S5 Amino acid sequence alignment of SlEIN4 and SlETR1‐SlETR7 in tomato. Black shade indicates amino acid residues 100% conserved, pink shade indicates amino acids that are >80% conserved, and blue shade indicates amino acids that are >50% conserved.
Figure S6 PCR genotyping of two T0 generation CR‐SlEIN4 mutants, PCR amplicon of Cas9, is a positive control.
Figure S7 Sequencing analysis of CR‐SlEIN4 alleles sequences from T2 homozygous lines of CR‐11 and CR‐13.
Figure S8 Fruit ripening phenotypes of WT, OE‐18 and CR‐11 plants. Scale bar, 2 cm.
Figure S9 Transverse section and quality of WT and transgenic fruits. (a) Transverse section of WT, OE‐18 and CR‐11 fruits. Scale bar, 10 mm. (b‐d) Fruit weight, sugar‐acid ratio and total soluble solids of WT and transgenic fruits at red ripe stage. Values are presented as means ± SD (n = 10). Asterisks indicate the significant difference between wild‐type and transgenic plants revealed by t‐test: *P < 0.05, **P < 0.01, ***P < 0.001.
Figure S10 Volcano map and clustering heatmap of differentially expressed genes (DEGs) between WT and CR lines. (a) Volcano map. The x‐coordinate represents the log2 (FoldChange), and the y‐coordinate represents the negative log value of 10 at the significance level. The two dotted lines in the figure represent the threshold values of the difference multiples, the dotted line is the threshold of significance level. The colour indicates whether the gene is up‐regulated, down‐regulated or insignificantly differentially expressed (DEGs were selected with a value of | log2 (FoldChange) | >1 and P adj < 0.05, as indicated below). (b) Clustering heatmap. Genes are represented horizontally, with one sample for each column. Red indicates high expression, and blue indicates low expression.
Figure S11 Sequencing of SlEIN4 haplotype in tomato varieties MT and M82.
Table S1 The geographic distribution of SNP (ch04_55126611) in 266 tomato accessions.
Table S2 List of 50 SNPs significantly associated with tomato fruit firmness.
Table S3 Genes included in the location of 55 127 814–55 157 228.
Table S4 Primers used for vector constructions.
Table S5 Primers used for positive and gene sequencing detection of transgenic plants.
Table S6 Primers used for qRT‐PCR.
Table S7 Reads quality test.
Table S8 Clean Reads comparison of reference genomes.
Table S9 Expression analysis of DEGs related to pectinase, ethylene or other hormone regulatory pathways.
Data Availability Statement
The data used to support the findings of this study are included in the article and the supporting information file.


