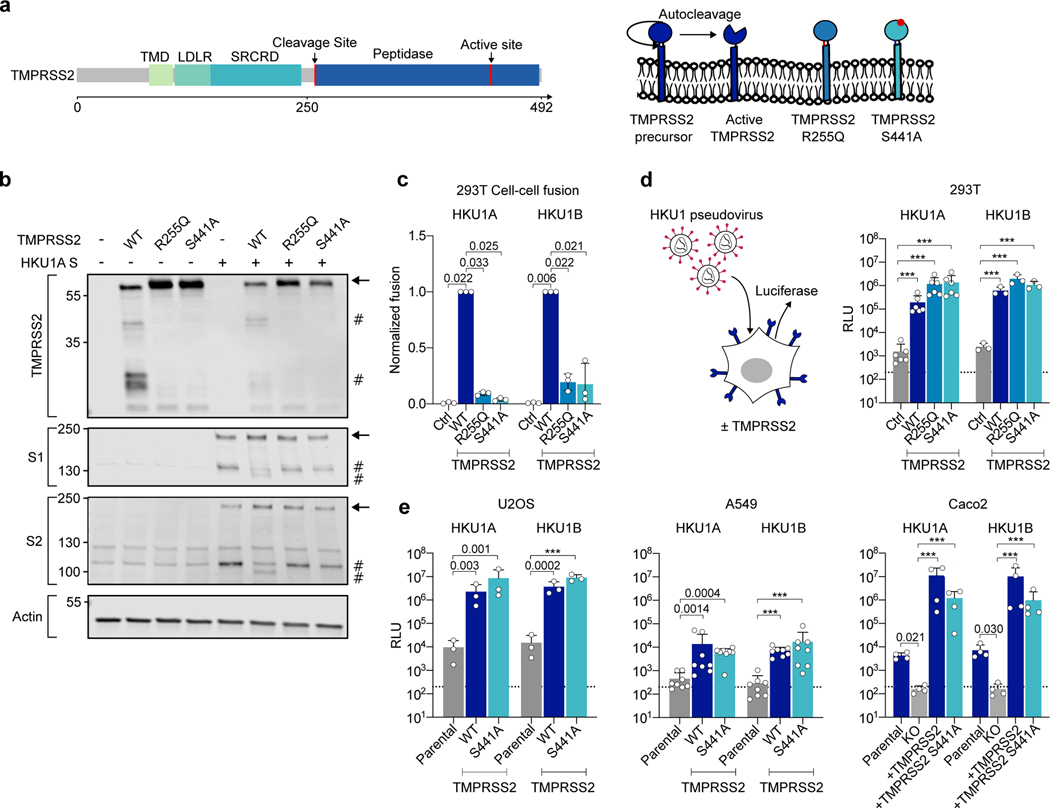
Figure 2. Effect of wild-type and mutant TMPRSS2 on HKU1 cell-cell fusion and pseudovirus infection.
a. TMPRSS2 protein. TMPRSS2 is composed of a Transmembrane Domain (TMD), a class A LDL receptor domain (LDLR), a Scavenger Receptor Cysteine-2 rich domain (SRCRD) and a serine peptidase. TMPRSS2 precursor autocleaves at R255-L256, resulting in an active protease. b. TMPRSS2 and spike cleavage. WB of 293T transfected with HKU1A spike and indicated TMPRSS2 mutants. One membrane was probed for S1, TMPRSS2 and actin, another for S2 and actin. For gel source data, see SI 1a, b. Representative blots of 3 independent experiments. Molecular weights: kDa. Arrows and # denote the uncleaved and cleaved proteins, respectively. c. Catalytically inactive TMPRSS2 mutants do not trigger HKU1 cell-cell fusion. Fusion of 293T-GFP split transfected with HKU1 spike and mutant TMPRSS2 was quantified after 20 h and normalized to WT TMPRSS2. Ctrl: pQCXIP-Empty. Left: HKU1A. Right: HKU1B. Data are mean ± SD of 3 independent experiments. d. Catalytically inactive TMPRSS2 mutants mediate HKU1 pseudovirus infection. 293T transfected with WT or mutant TMPRSS2 were infected by Luc-encoding HKU1 pseudovirus. Luminescence was read 48 h post infection. Dotted line indicates background in non-infected cells. Ctrl: pQCXIP-Empty. Data are mean ± SD of 6 (HKU1A) or 3 (HKU1B) independent experiments. e. HKU1 pseudovirus infection in cell lines stably expressing TMPRSS2. Left: U2OS cells. Middle: A549 cells. Right: Caco2 cells. The TMPRSS2 KO Caco2 cells were stably transduced with indicated TMPRSS2. Data are mean ± SD of 3 (U2OS), 8 (A549), 4 (Caco2) independent experiments. Statistical analysis: c: one-way ANOVA on non-normalized data with Dunnett’s multi-comparison test compared to WT TMPRSS2 expressing cells. d, e: one-way ANOVA on log-transformed data with Dunnett’s multi-comparison test compared to the untransfected (d), parental (e: U2OS and A549) or KO cells (e: Caco2), ***p<0.0001.