Fig. 2. ATF4 ablation disrupts expression of genes involved in amino acid and one-carbon metabolism.
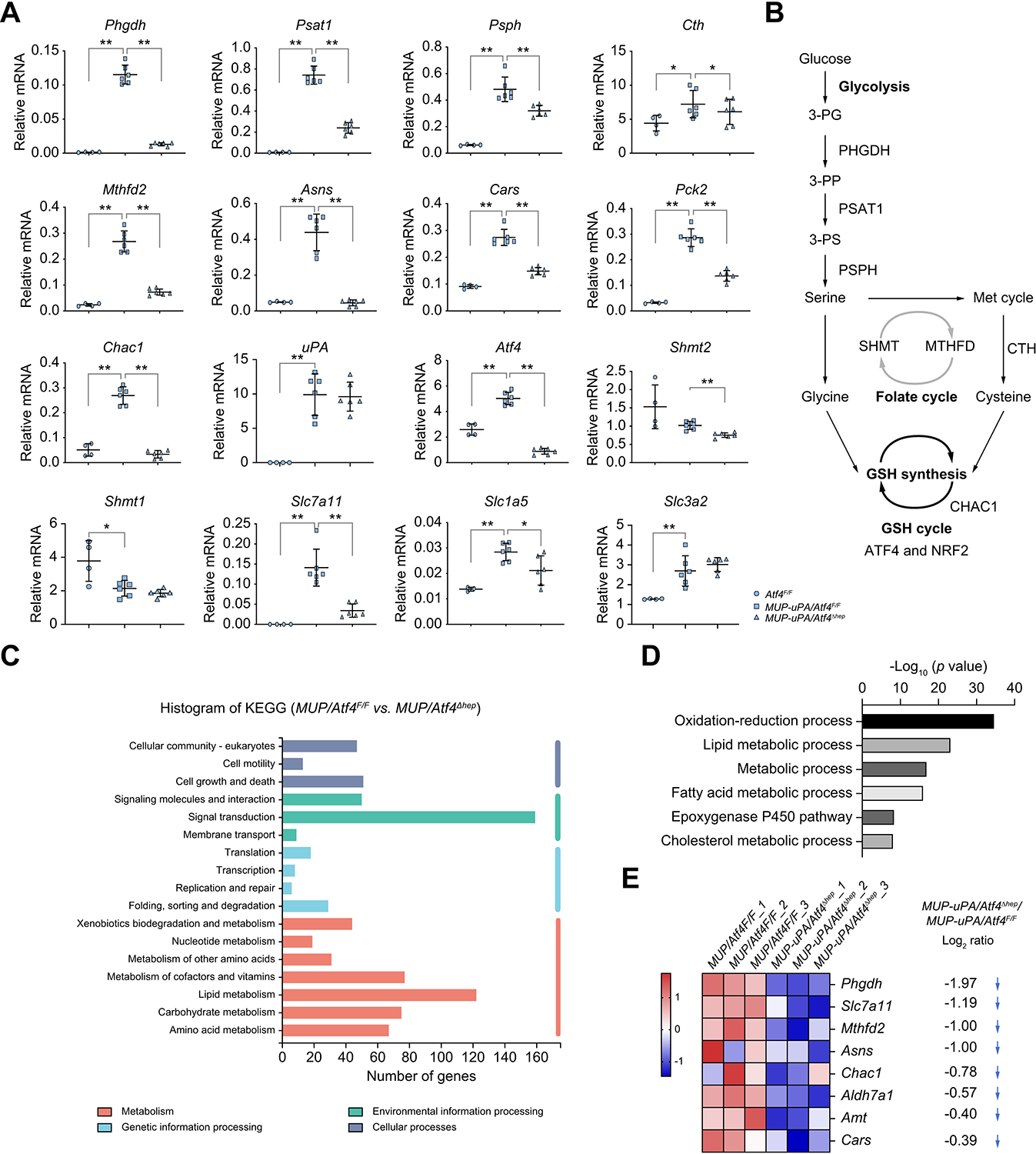
(A) qRT-PCR analysis of liver mRNAs from 6-wo Atf4F/F, MUP-uPA/Atf4F/F, and MUP-uPA/Atf4Δhep mice. Mean ± SD (n = 3–6/group). *p <0.05; **p <0.01 (Student’s t test). (B) Diagram showing metabolic pathways with key enzymes involved in serine, glycine, cysteine, and GSH metabolism as well as the folate and methionine cycles. (C) KEGG pathway analysis of genes differentially represented in RNA-seq data from livers of overnight fasted 6-wo MUP-uPA/Atf4Δhep and MUP-uPA/Atf4F/F mice (n = 3). (D) DAVID-based GO analysis of differentially expressed genes in above RNA-seq data (n = 3). (E) Heat map representation of significantly downregulated genes in MUP-uPA/Atf4Δhep relative to MUP-uPA/Atf4F/F livers (n = 3) associated with ‘amino acid metabolism’ including Log2FC value. ATF4, activating transcription factor 4; CHAC1, glutathione specific gamma-glutamylcyclotransferase 1; CTH, cystathionine gamma-lyase; GO, gene ontology; GSH, glutathione; KEGG, Kyoto Encyclopedia of Genes and Genomes; Log2FC, log2 fold-change; MTHFD2, methenyltetrahydrofolate dehydrogenase 2; MUP-uPA, major urinary protein promoter–urokinase plasminogen activator; NRF2, nuclear factor erythroid 2-related factor 2; PHGDH, phosphoglycerate dehydrogenase; PSAT1, phosphoserine aminotransferase 1; PSPH, phosphoserine phosphatase; qRT-PCR, quantitative reverse-transcription PCR; RNA-seq, RNA sequencing; SHMT, serine hydroxymethyltransferase; 3-PG, 3-phosphoglycerate; 3-PP, 3-phosphohydroxypyruvate; 3-PS, 3-phosphoserine; wo, week old.
