Extended Data Fig. 6 |. Mechanism of TIR tetramerization.
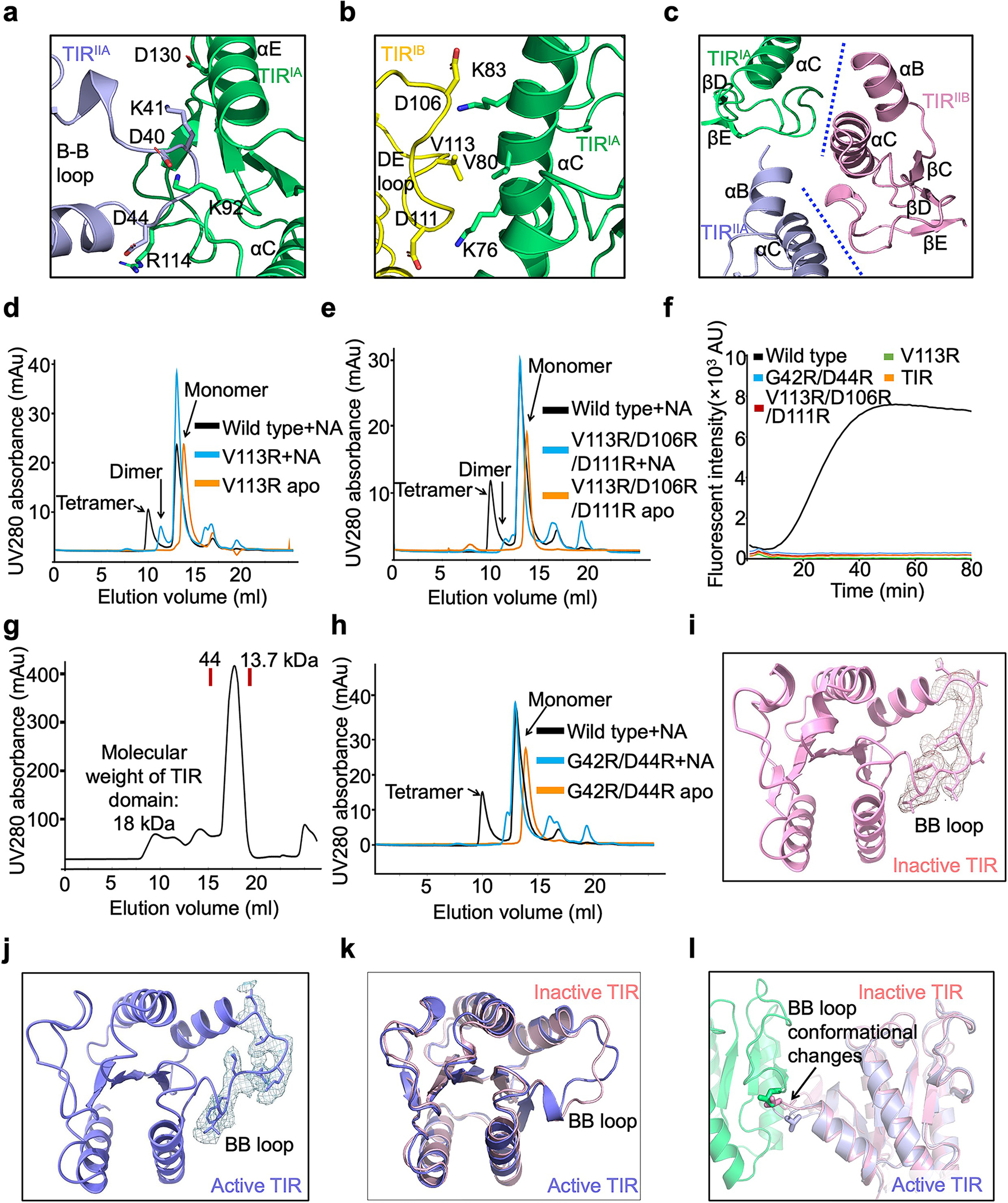
a, Detailed interactions between TIRIA and TIRIIA with interfacial residues in sticks.
b, Interface between TIR and TIR with key residues highlighted in sticks.
c, TIRIIB engages with TIRIA and TIRIIA via tetramerization interfaces.
d, Compared to wild type, V113R eluted as dimers and monomers in the presence of RNA/DNA.
e, Compared to wild type, V113R/D106R/D111R eluted as dimers and monomers in the presence of RNA/DNA.
f, Representative kinetic data of NAD+ hydrolysis by wild type and mutant MapSPARTA.
g, Gel filtration profile of TIR domain alone, showing that TIR domain eluted as a monomer.
h, Compared to wild type, G42R/D44R eluted as monomers in the presence of RNA/DNA.
i, j, BB loop in TIR in inactive state (i) and active state (j) fitted into cryo-EM densities at 2.0 𝛔.
k, Overlaid structures of TIR in inactive state (pink) and in active state (blue), revealing conformational changes of the BB-loop.
l, BB loop conformational changes are critical for the formation of the asymmetric dimer. Inactive TIR modelled into the asymmetric dimer revealed that the BB loop in inactive state could clash with the other protomer.
