Extended Data Fig. 9 |. Recognition of RNA by MapSPARTA.
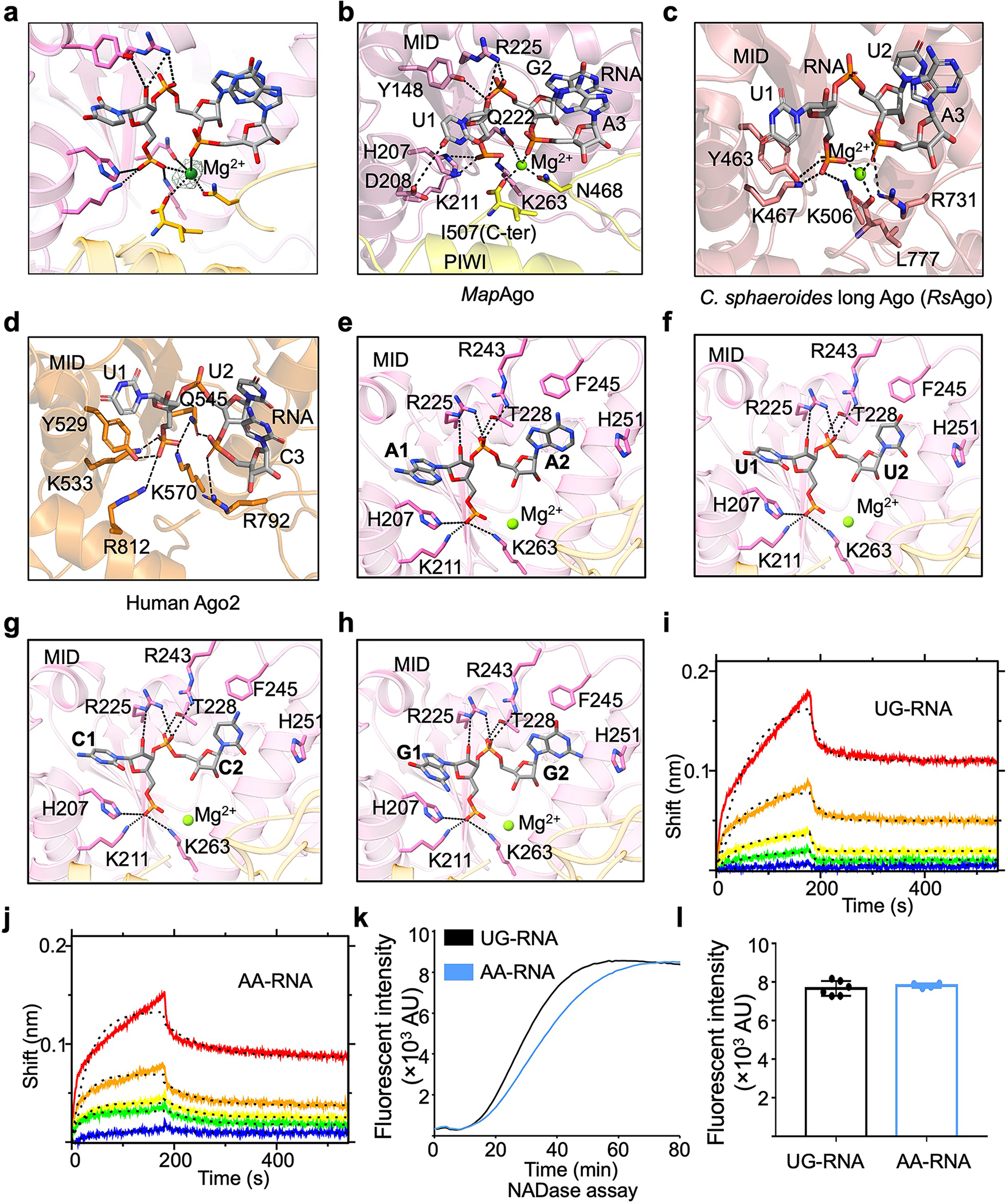
a, A Magnesium ion fitted into cryo-EM density at 2.5 𝛔.
b-d, Expanded views of the 5’ nucleotide of guide RNA coordinated by residues in pockets of MID domains from MapAgo (b), Cereibacter sphaeroides long Ago (PDB ID 5AWH, c), and human Ago2 (PDB ID 4W5T, d). Magnesium ions in spheres are responsible in coordinating guide RNA by interacting with phosphate groups. Residues involved in coordinating guide RNA are highlighted in sticks.
e-h, AA (e), UU (f), CC (g), and GG (h) as the first and second RNA nucleotides were modelled into the binding pocket, revealing a similar mechanism of being recognized by MapSPARTA.
i, j, The sensorgrams of wild type SPARTA binding to the chip-immobilized RNAs, UG-RNA (i) and AA-RNA (j), are expressed in shift versus time. The protein concentrations were 50, 25, 12.5, 6.25, and 3.12 nM (from top to bottom).
k, Representative kinetic data of NAD+ hydrolysis by MapSPARTA in the presence of UG-RNA or AA-RNA.
l, Plotted graphs of MapSPARTA catalysis stimulated by UG-RNA or AA-RNA. Data are mean ± SD from more than 3 replicates as indicated (UG-RNA, n=6; AA-RNA, n=4).
