Fig. 4 |. Recognition of nucleic acid.
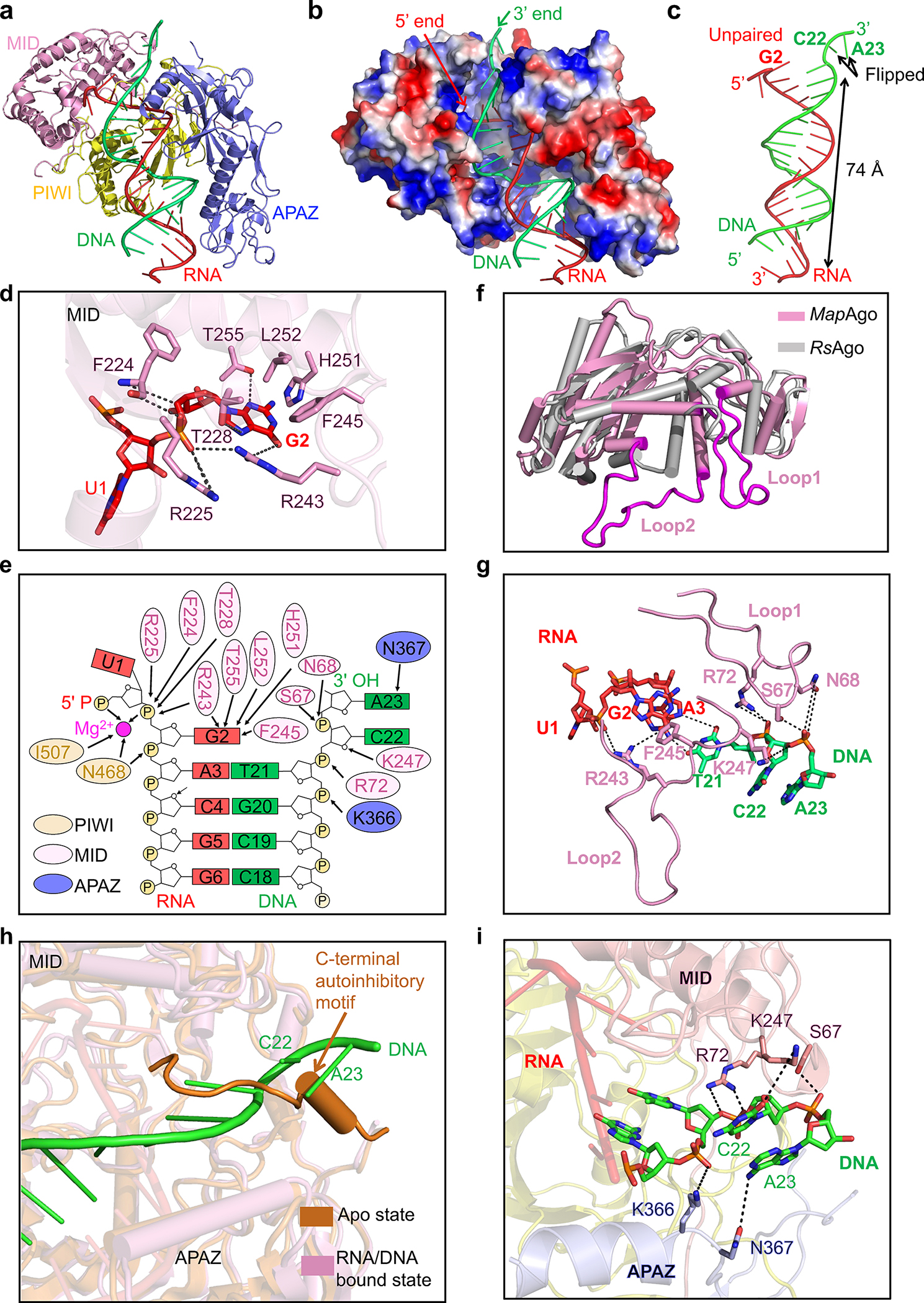
a, Ribbon diagram of MapSPARTA bound to guide RNA (red) and target DNA (green). TIR is omitted because of its lack of interaction with nucleic acids.
b, Electrostatic surface representation of MapSPARTA in complex with the RNA-DNA duplex.
c, Structure of the guide RNA-target DNA duplex.
d, Detailed interactions between the G2 nucleotide of guide RNA and the MID domain of pAgo, with key residues shown as sticks.
e, Coordination of G2 of guide RNA and C22 and A23 of target DNA by MapSPARTA. Residues from PIWI domain, MID domain and APAZ domain are colored in yellow, pink, and blue, respectively.
f, Structural comparison of MapAgo (Ago of MapSPARTA) and Cereibacter sphaeroides Ago (RsAgo), revealing the two long loops unique to MapAgo.
g, Lack of Watson-Crick pairing between RNA and DNA due to steric interference by the two long loops of pAgo. Key residues in the long loops are shown as sticks.
h, Overlaid structures of MapSPARTA in apo monomer and RNA/DNA-bound tetramer states, revealing the clash of DNA with the CTM of APAZ in the apo state.
i, Detailed interactions of the DNA 3’ end with the MID domain of pAgo (pink) and the APAZ domain of APAZ-TIR (blue). Key residues coordinating DNA are shown as sticks.
