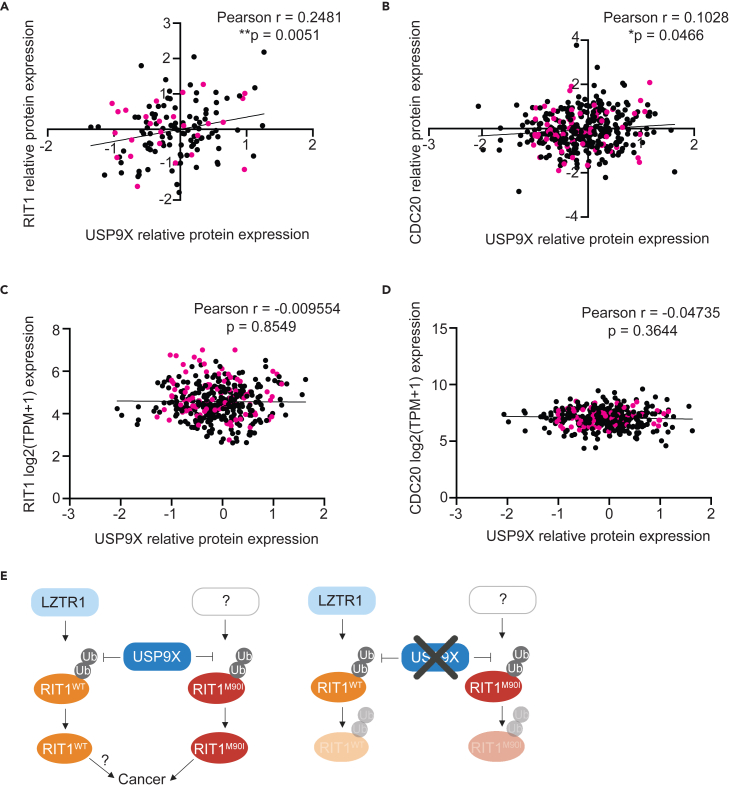
Figure 6.
USP9X-mediated regulation of RIT1 is relevant across cancer types
(A) Correlation of proteomics data50 from the Cancer Dependency Map (DepMap) comparing USP9X (Q93008) and RIT1 (Q92963-3). Pearson r and p values calculated in Prism. Pink dots indicate non-small cell lung cancer cell lines.
(B) Correlation of DepMap proteomics data50 comparing USP9X (Q93008) and CDC20 (Q12834). Pearson r and p values calculated in Prism. Pink dots indicate non-small cell lung cancer cell lines.
(C) Correlation of DepMap mRNA expression data (Expression Public 23Q2)51 for RIT1 and USP9X proteomics data (Q93008).50 Pearson r and p values calculated in Prism. Pink dots indicate non-small cell lung cancer cell lines.
(D) Correlation of DepMap mRNA expression data (Expression Public 23Q2)51 for CDC20 and USP9X proteomics data (Q93008).50 Pearson r and p values calculated in Prism. Pink dots indicate non-small cell lung cancer cell lines.
(E) Proposed model (left) of RIT1 protein regulation. RIT1WT is ubiquitinated by LZTR1, while RIT1M90I is ubiquitinated by a currently unknown E3 ligase. USP9X counteracts the ubiquitination of both wild-type and mutant RIT1. Increased RIT1 abundance and stability are important for RIT1 function and disease progression. The exact biological consequences of RIT1WT amplification have yet to be elucidated. Genetic knockout (right) of USP9X prevents RIT1 deubiquitination, thereby promoting RIT1 degradation and abrogating oncogenic phenotypes. Figure created with BioRender.com.
See also Figure S5.