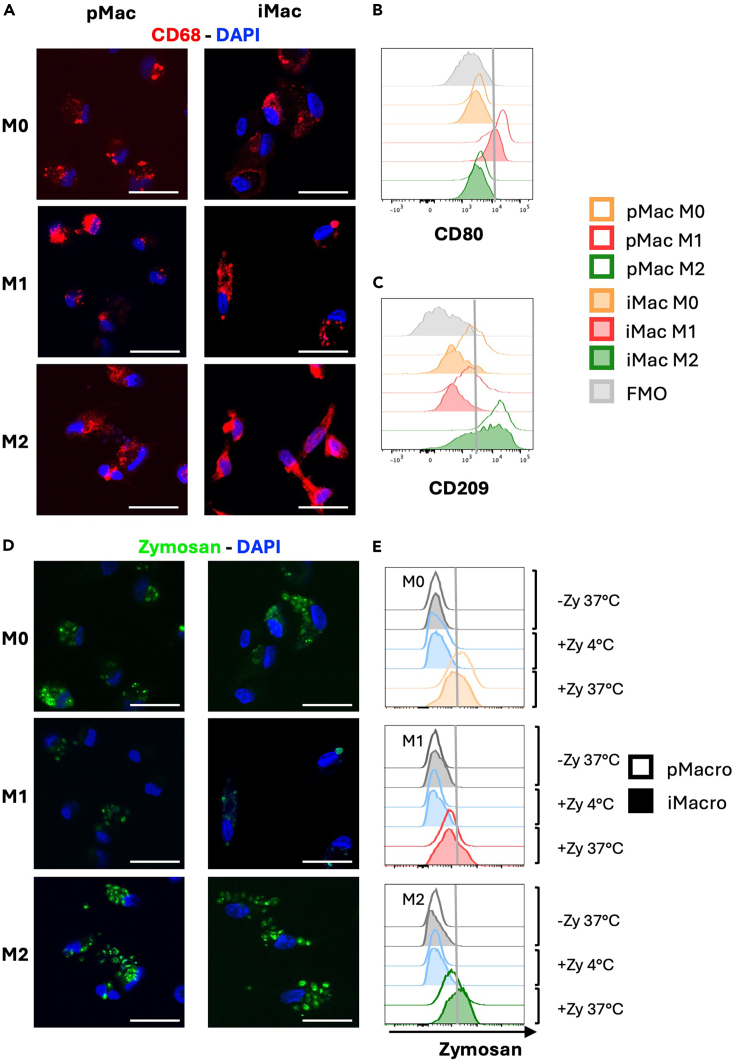
Figure 6.
Phenotypic and functional assessment of hiPSC-derived and primary macrophages
(A) Representative images of immunofluorescence CD68 staining observed in primary macrophages (pMac, n = 5; first column) and iPSC-derived macrophages (iMac, n = 3; second column) polarized in M0, M1, and M2 macrophages (First, second and third row, respectively). Cells were stained for CD68 (red) and with DAPI for nuclear staining (blue).
(B and C) Representative overlay showing the expression of the M1 marker CD80 (B) and the M2 marker CD209 (C) on primary macrophages (pMac, n = 4; empty histograms) and iPSC-derived macrophages (iMac, n = 3; filled histograms) polarized in M0 (orange), M1 (red), and M2 (green) macrophages. The fluorescence minus one (FMO) used as negative control is shown (gray, filled histogram). (D-E) Phagocytic ability of M0, M1 and M2 primary macrophages (pMac) and iPSC-derived macrophages (iMac) was evaluated by using pHrodo Green Zymosan A Bioparticles and was assessed by flow cytometry and confocal microscopy.
(D) Representative images showing phagocytized Zymosan particles (in green) in primary macrophages (pMac, n = 3; first column) and iMac (iMac-Con, n = 3; second column) polarized in M0, M1, and M2 macrophages (First, second and third row, respectively). Nuclei were stained with DAPI (blue).
(E) Representative overlay showing the Zymosan phagocytosis in primary macrophages (pMac, n = 4; empty histograms) and iPSC-derived macrophages (iMac, n = 3; filled histograms) polarized in M0 (in orange, first graph), M1 (in red, second graph), and M2 (in green, third graph) macrophages. Samples incubated without Zymosan (gray histograms) and with Zymosan incubated at 4°C (light blue histograms), which were used as negative controls, are shown.
Scale bars = 25 μm and are applicable to all images.