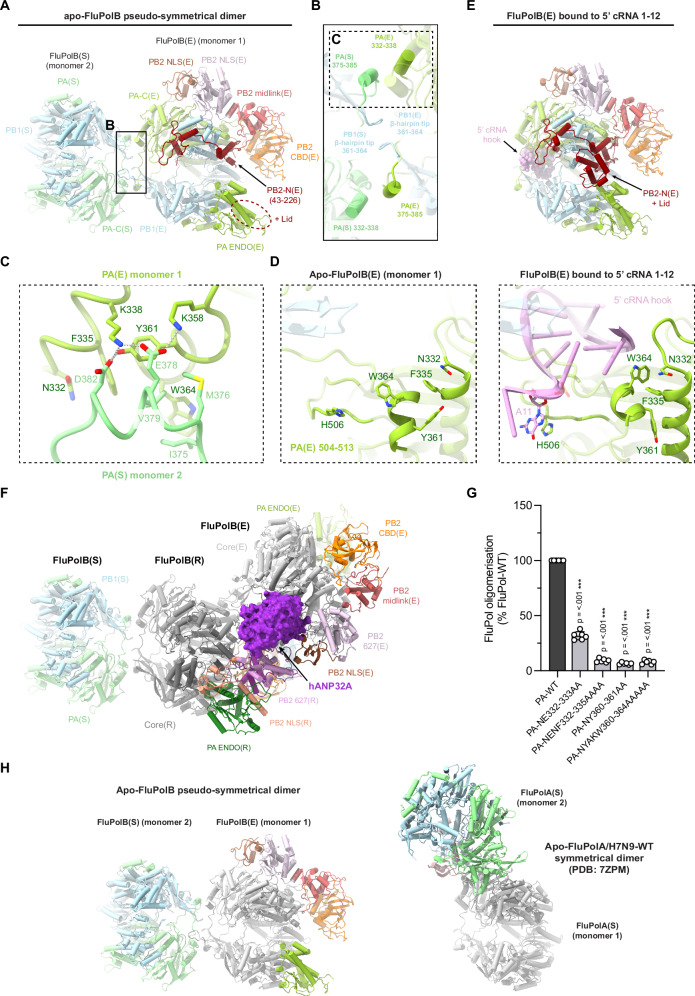
Fig. 4. Apo-FluPolB pseudo-symmetrical dimer and 5’ cRNA bound FluPolB encapsidase.
A Cartoon representation of the most abundant apo-FluPolB pseudo-symmetrical dimer. Monomer 1 has the encapsidase conformation, FluPolB(E) with PA-C(E) in light green, PB1(E) in light blue, PB2-N(E) (43-226) in dark red, PB2 midlink(E) in salmon, PB2 CBD(E) in orange, PB2 627(E) in light pink, PB2 NLS(E) in brown. Putative PB2-N(E) lid density (dotted ellipse) is located next to PA-ENDO(E). Monomer 2, FluPol(S), takes multiple conformation. Here, only PA(S) and PB1(S) subunits are shown, respectively in light green and light blue. The cores from each monomer form a symmetrical interface highlighted by a dotted rectangle, corresponding to (B). B Close-up view of the FluPolB symmetrical dimer interface. The main interaction is mediated by PA(E)/332-338 interacting with PA(S)/375-385. PB1 β-hairpin tips from both monomers interact with each other across the 2-fold axis. One of the symmetrical dimer interfaces is highlighted with a dotted rectangle, corresponding to (C). C Detail of the residue contacts between PA(E)/332-338 and PA(S)/375-385 at the symmetrical dimer interface. Domains are coloured as in (A, B). Ionic and hydrogen bonds are shown as grey dotted lines. D Structural rearrangement of PA(E) upon 5’ cRNA hook binding. The 5’ cRNA hook (nts. 1–12) is coloured plum with nucleotides as stubs. E Cartoon representation of the 5’ cRNA bound FluPolB encapsidase structure. Domains are coloured as in (A). The 5’ cRNA hook (nts. 1–12) is displayed as spheres and coloured plum. The PB2-N(E) lid is observed when FluPol(E) is bound to the 5’ cRNA hook. F Cartoon representation of the complete FluPolB trimer, composed of the replication complex (FluPolB(R)+hANP32A+FluPolB(E)), with FluPolB(R) core forming a pseudo-symmetrical dimer with a third FluPolB(S). FluPolB(S) is orientated and coloured as in (A). hANP32A is displayed as a purple surface. FluPolB(R) core is dark grey, PA ENDO(R) is dark green, PB2 midlink(R) is magenta, PB2 CBD(R) is orange, PB2 627(R) is pink and PB2 NLS(R) is beige. FluPolB(E) core is light grey, PA ENDO(E) is light green, PB2 midlink(E) is salmon, PB2 CBD(E) is orange, PB2 627(E) is light pink and PB2 NLS(E) is brown. G Cell-based split-luciferase complementation assay to assess B/Memphis/13/2003 FluPol self-oligomerisation for the indicated PA mutants. HEK-293T cells were co-transfected with plasmids encoding PB2, PA, PB1-luc1 and PB1-luc232. Luminescence signals due to luciferase reconstitution are represented as a percentage of PA-WT (mean ± SD, n = 6, ***p < 0.001, one-way ANOVA; Dunnett’s multiple comparisons test). Source data are provided as a Source Data file. H Comparison of the distinct apo-FluPolB (left) and apo-FluPolA/H7N9-WT (PDB 7ZPM) (right) symmetrical dimers, with monomer 1 core (grey) in the same orientation. Other domains are coloured as in (A).