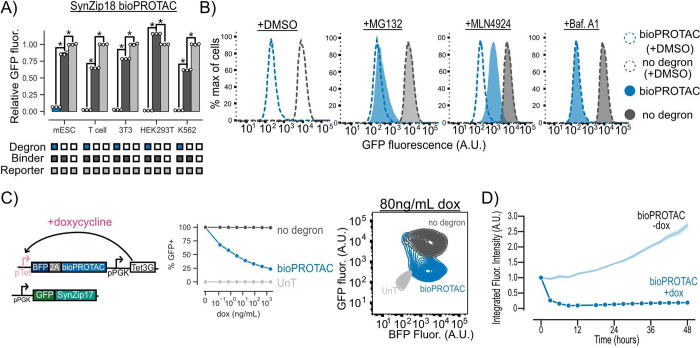
Figure 2.
bioPROTACs are versatile tools for protein degradation capable of dose-dependent and rapid degradation of proteins. (A) BioPROTACs are capable of potent degradation across a variety of mammalian cell types. GFP fluorescence was measured by flow cytometry, and “Relative GFP fluorescence” was quantified as described in Figure 1 for each cell type. Each dot represents a technical replicate, and error bars show SEM. (B) bioPROTAC degradation of cytosolic proteins relies on the proteasome via cullin ring ligases. SynZip18 bioPROTAC and GFP reporter expressing Jurkat T cells or control lines were treated with either MG132, Bafilomycin A1, MLN4924, or a DMSO control. GFP fluorescence was then measured by flow cytometry. Histograms are representative of three biological replicates. (C) bioPROTAC titration results in dose-dependent degradation of cytosolic proteins. Left: Cartoon depicting lentiviral payloads encoding a GFP reporter protein and a doxycycline inducible bioPROTAC and the Tet3G protein. Middle: After isolation by FACS, cells were treated with a 5-fold titration series of doxycycline or a media only control for 48 h. GFP fluorescence was measured by flow cytometry, and GFP+ cells were selected relative to an untransduced control. Each dot represents the mean of three biological replicates. Error shows SEM. Right: Representative contour plot of bioPROTAC expression level (BFP) and GFP expression level at 80 ng/mL of doxycycline. (D) bioPROTACs induced substantial GFP loss after 4 h of doxycycline treatment. The doxycycline inducible Jurkat T cell line described above was treated with either 2000 ng/mL doxycycline or a media only control. GFP fluorescence was measured by live cell imaging and analyzed using the Incucyte software. Each trace is normalized to integrated fluorescent intensity values at the initial measurement time point. Each dot is the mean of three biological replicates. Error shows SEM. An unpaired t test was used for all statistical comparisons. *P < 0.01