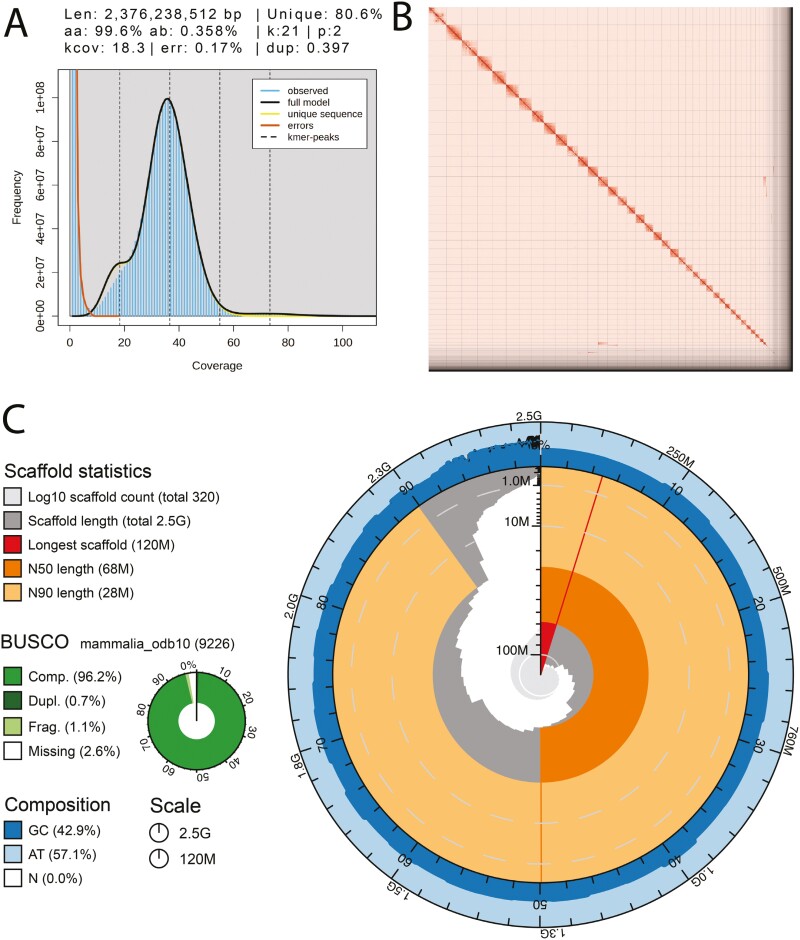
Fig. 2.
Visualization of assembly metrics. (A) K-mer frequencies from the adapter-trimmed PacBio HiFi data used to estimate genome size, sequencing error rate, and heterozygosity. The main peak at ~37× coverage corresponds to homozygous regions of the genome, while the slight peak at ~18× corresponds to heterozygous regions of the genome. The peak around zero corresponds to probable sequencing errors. (B) The omni-C contact map for the primary assembly after manual curation shows the 3D organization of the genome, with darker areas indicating closer proximity. (C) Snail plot displaying assembly metrics for the primary assembly.