Fig 5.
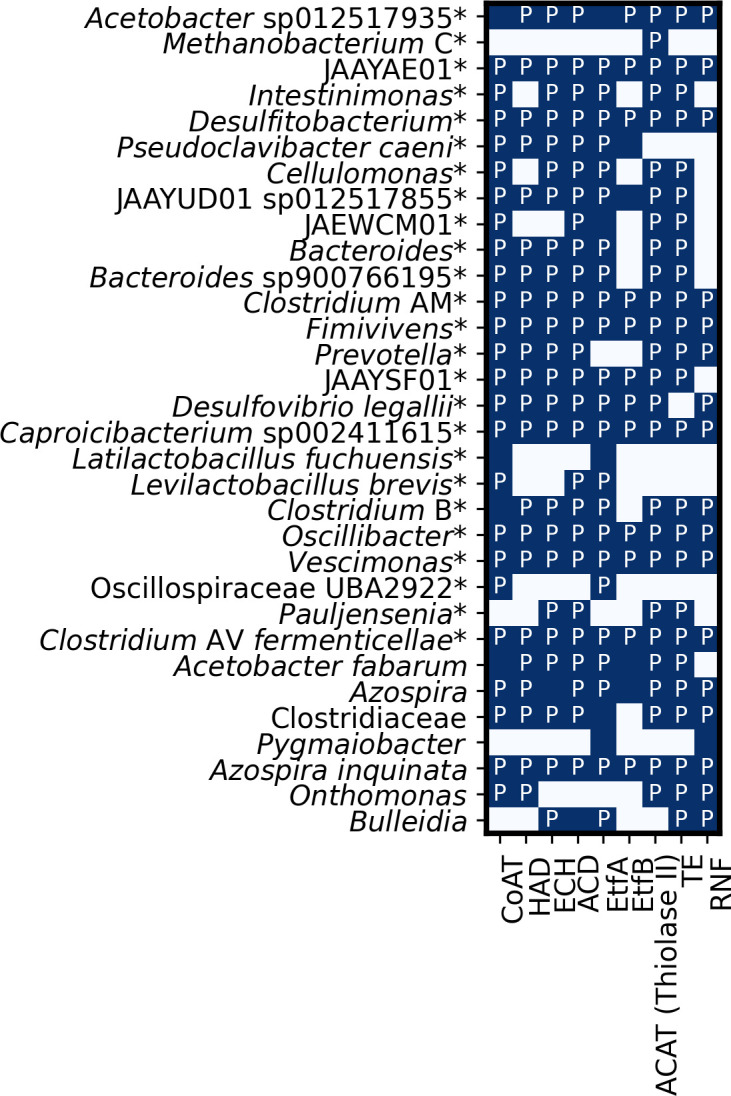
Absence or presence of nine enzymes involved in the RBOX pathway (acronyms described previously) in reactor de-novo assembled metagenomes and proteomes as monitored by shotgun metagenomics analysis and proteomics. MAG taxonomy was assigned using gtdb-tk (r220). Enzyme acronyms were described in Fig. 1. A blue box denotes the presence in the metagenome, the letter P the presence in the metaproteome, and a white box without a P the absence in both the metagenome and metaproteome. MAGs identified to species level are depicted. A * indicates high-quality MAGs >90% complete and <5% contaminated (determined with CheckM).
