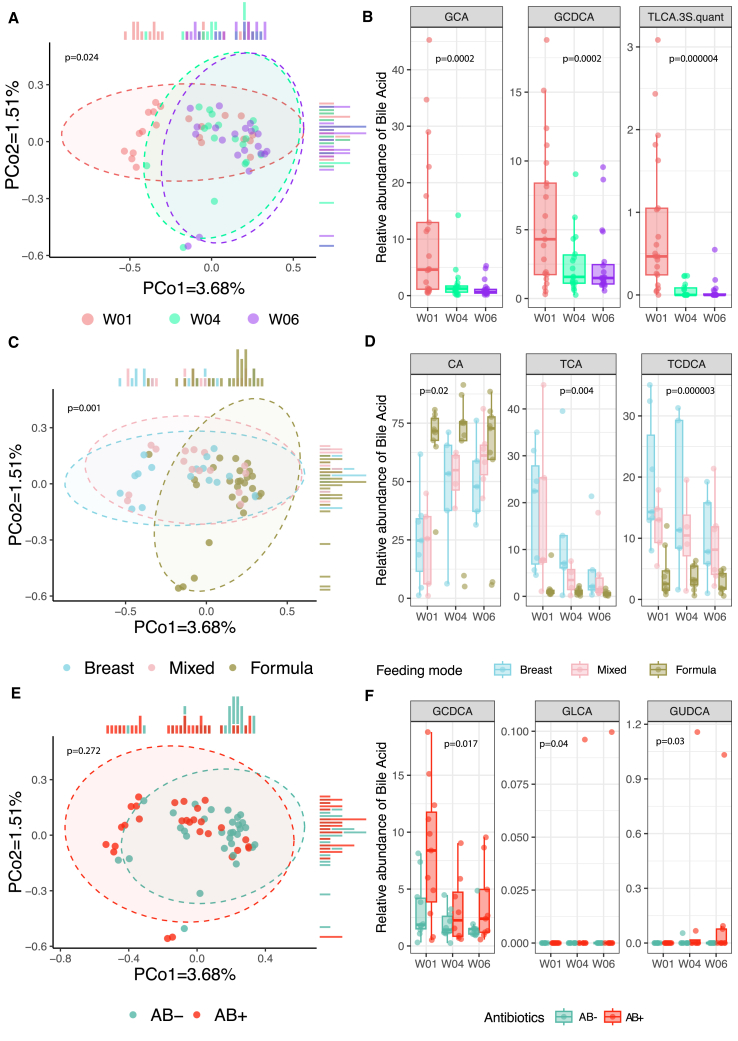
Figure 3.
Infant fecal bile acids in relation to time point, feeding mode, and antibiotic groups
(A) PCoA plot using Canberra distances. Dot color indicates the time point. p value was derived from a PERMANOVA test with 999 permutations without correction for other phenotypes.
(B) Boxplots of the relative abundance of the most significantly associated bile acids with time. p value derived from a linear mixed-effect model without correction for other phenotypes. GCA, glycocholic acid; GCDCA, glycochenodeoxycholic acid; TLCA.3S.quant, taurolithocholic acid 3-sulfate.
(C) PCoA plot using Canberra distances. Dot color indicates the feeding mode of each infant at each time point. p values derived from a PERMANOVA test with 999 permutations without correction for other phenotypes.
(D) Boxplots of the relative abundance of the most significantly associated bile acids with feeding mode (Table S6B). p value derived from a linear mixed-effect model without correction for other phenotypes. CA, cholic acid; TCA, taurocholic acid; TCDCA, taurochenodeoxycholic acid.
(E) PCoA plot using Canberra distances. Dot color indicates the antibiotic group the infant belongs to (AB+ or AB−). p values derived from a PERMANOVA test with 999 permutations after correction for feeding mode.
(F) Boxplots of the relative abundance of the most significantly associated bile acids with AB group after correction for feeding mode (Table S6C). GCDCA, glycochenodeoxycholic acid; GLCA, glycolithocholic acid; GUDCA, glycoursodeoxycholic acid. In (A)–(F), each dot represents a sample, with the following sample sizes: W01 (n = 21), W04 (n = 18), and W06 (n = 20) from a total of 22 unique infants. Boxplots visualize the median, hinges (25th and 75th percentiles), and whiskers extending up to 1.5 times the interquartile range from the hinges.