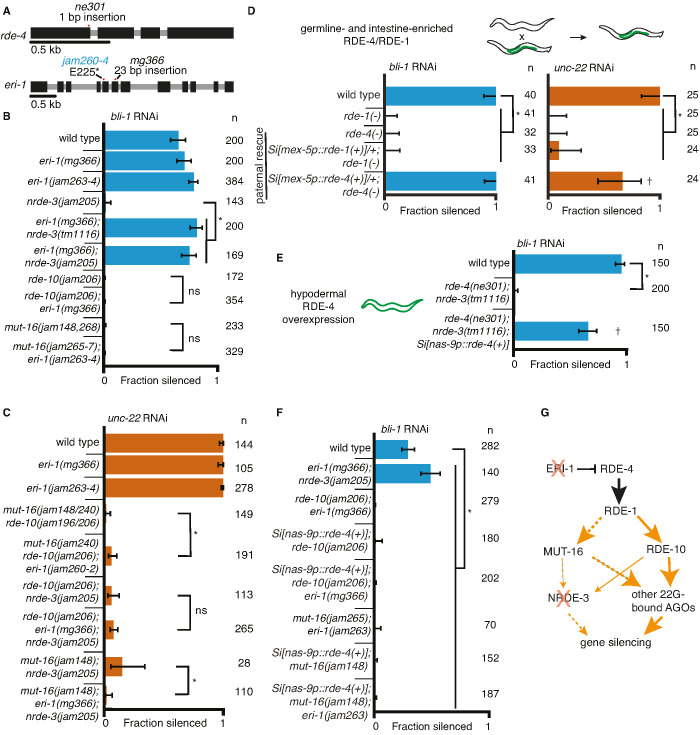
Figure 3. Gene-specific requirements for NRDE-3 can be bypassed in two ways.
(A) Gene schematics (as in Figure 1) of rde-4 and eri-1. (B, C) Loss of ERI-1 can bypass the NRDE-3 requirements for silencing bli-1 but not the other requirements for silencing bli-1 or unc-22. Feeding RNA interference (RNAi) targeting bli-1 (B) or unc-22 (C) with fractions silenced, numbers scored, comparisons, asterisks, and error bars as in Figure 1. (B) Loss of ERI-1 (mg366, jam263, and jam264 alleles) can compensate for the role of NRDE-3 (tm1116 and jam205 alleles) but not of RDE-10 (jam206 allele) or MUT-16 (jam148, jam265, jam266, jam267, and jam268 alleles) in bli-1 silencing. See Table 3 for additional information. (C) Silencing of unc-22 is not restored by loss of ERI-1 (mg366, jam260, jam261, and jam262 alleles) in mutants that also lack any two of mut-16 (jam148 and jam240 alleles), rde-10 (jam196 and jam206 alleles), or nrde-3 (jam205 allele). See Table 3 for additional information. (D, E) Overexpression of RDE-4 in the hypodermis can bypass the requirement for NRDE-3 in bli-1 silencing. (D) Minimal amounts of RDE-4 are sufficient for somatic silencing. (Top) Schematic depicting generation of male progeny with paternal inheritance of a single-copy transgene (Si[…]) that expresses rde-4(+) or rde-1(+) under the control of the mex-5 promoter (mex-5p) in the germline (green) of rde-4(−) or rde-1(−) animals, respectively (germline- and intestine-enriched RDE, based on rescue of RNAi in rde-1(−) animals (39)). (Bottom) Male cross progeny with the transgene were scored after feeding only F1 animals, showing that unlike animals with germline- and intestine-enriched RDE-1, animals with similarly enriched RDE-4 can rescue both unc-22 and bli-1 silencing. Thus, small amounts of RDE-4 potentially mis-expressed in the hypodermis or a non-autonomous effect of RDE-4 from the germline or intestine is sufficient for silencing in the muscle and hypodermis. † indicates p < 0.05 when compared to either wild type or the rde-4(−) mutant and other symbols are as in (B). (E) Silencing of bli-1 is restored in nrde-3(tm1116); rde-4(ne301) double mutants when rde-4(+) is overexpressed in the hypodermis (Si[nas-9p::rde-4(+)]). (F) Silencing of bli-1 is not restored in animals lacking MUT-16 or RDE-10, despite the overexpression of RDE-4 in the hypodermis and/or the loss of ERI-1. (G) Summary depicting the bypass of NRDE-3 when ERI-1 is eliminated and/or RDE-4 is overexpressed. The increase in double-stranded RNA (dsRNA) processing increases the contributions of NRDE-3-independent paths to silencing.
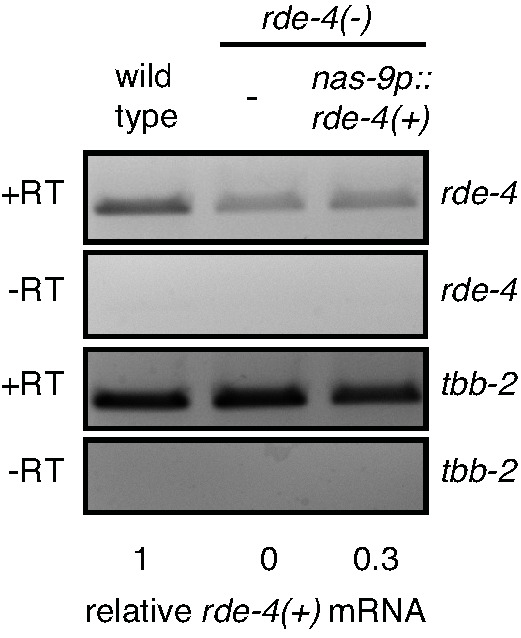
Figure 3—figure supplement 1. Overexpressing RDE-4 in the hypodermis.