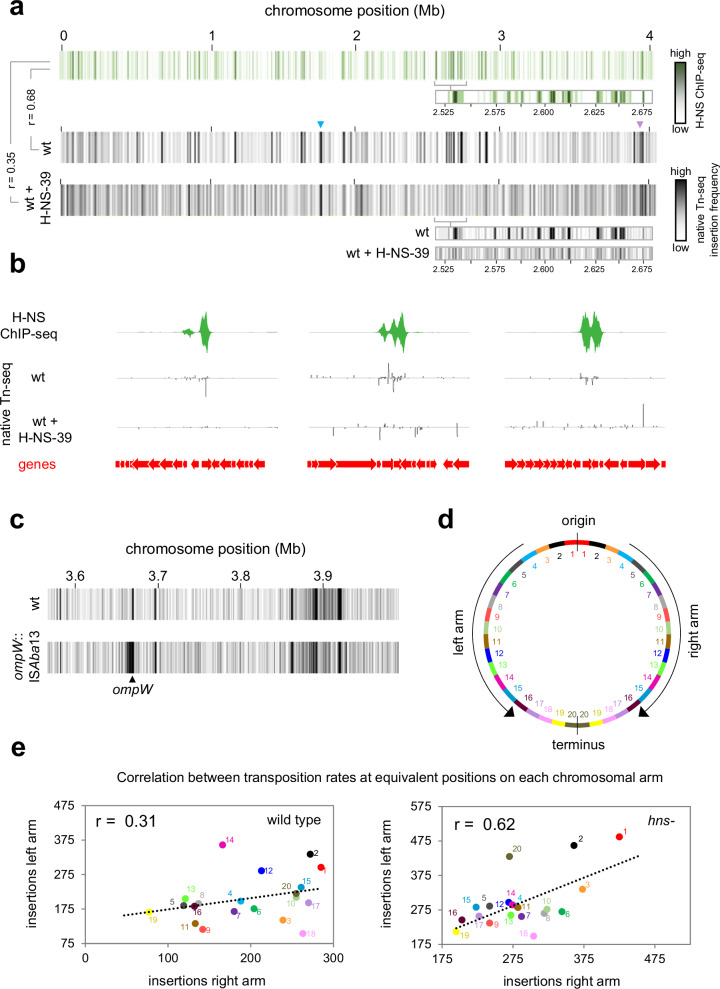
Fig. 5. DNA bridging is required for capture of transposition events by H-NS.
a H-NS-39 causes more uniform ISAba13 transposition patterns globally. The panel shows three heatmaps, each representing the A. baumannii chromosome divided into 1 kb sections. Sections are coloured according to the H-NS ChIP-seq binding signal (top) or the number of ISAba13 insertions detected using native Tn-seq in the absence (middle) or presence (bottom) of H-NS-39 expression. The heatmap expansions are provided to aid comparison of H-NS binding and insertion frequency. Pearson correlation coefficients (r) between datasets are shown. b Examples of H-NS-39 mediated changes to ISAba13 transposition patterns. Selected chromosomal regions with H-NS ChIP-seq and native Tn-seq data shown. In both cases, traces indicate read depths with positive and negative values corresponding to the top and bottom DNA strand respectively. Genes are shown as arrows. c Transposition hotspots arise immediately adjacent to existing copies of ISAba13. The panel shows two heatmaps, each representing a 0.4 Mb region of the A. baumannii chromosome divided into 1 kb sections. Introduction of ISAba13 in ompW creates a transposition hotspot, around 10 kb in size, surrounding the insertion site. d Map of the A. baumannii chromosome. The schematic illustrates the A. baumannii chromosome divided into 40 sections, each 100 kb in length. Equivalent sections in the left and right replichores are coloured and numbered accordingly. e The number of detected transposition events correlate between equivalent positions on each chromosomal arm. The scatter plots show the number of ISAba13 insertions, detected by native Tn-seq, at equivalent positions on each chromosomal arm, in the presence and absence of hns. Data points are coloured according to the schematic in (d) and r values are correlation coefficients. Source data are provided as a Source Data file.