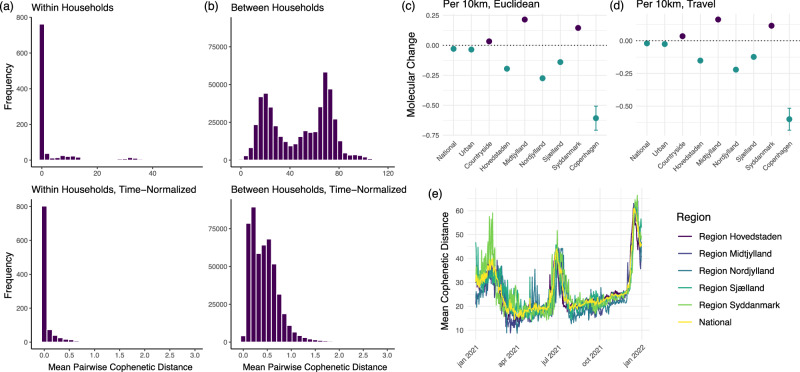
Fig. 6. Relationship between geographic and genomic distances alongside mean cophenetic distances within and between households, and mean cophenetic distance over time by region.
a Distribution of mean pairwise cophenetic distances between individuals within the same household (n = 1000 households), normalised to time (i.e. distance divided by time in days between individuals testing positive). b Distribution of mean pairwise cophenetic distances between individuals in different households (n = 1000 households), normalised to time. c Molecular change (i.e. number of nucleotide changes) per 10 km increase in Euclidean distance across various geographic models (national, residential zone, regional, city) (National: n = 20,000 individuals; Urban: n = 18,111; Countryside: n = 1817; Hovedstaden: n = 1000; Midtjylland: n = 1000; Nordjylland: n = 1000; Sjælland: n = 1000; Syddanmark: n = 1000; Copenhagen: n = 3416). Error bars denote 95% confidence intervals. d Molecular change per 10 km increase in car travel distance using OpenStreetMap across different geographic models (national, residential zone, regional, city) (National: n = 20,000 individuals; Urban: n = 18,111; Countryside: n = 1817; Hovedstaden: n = 1000; Midtjylland: n = 1000; Nordjylland: n = 1000; Sjælland: n = 1000; Syddanmark: n = 1000; Copenhagen: n = 3416). Error bars denote 95% confidence intervals. e Mean pairwise cophenetic distance over time, stratified by region (n = 10,000 individuals per region, n = 20,000 for the national subset).