Figure 3.
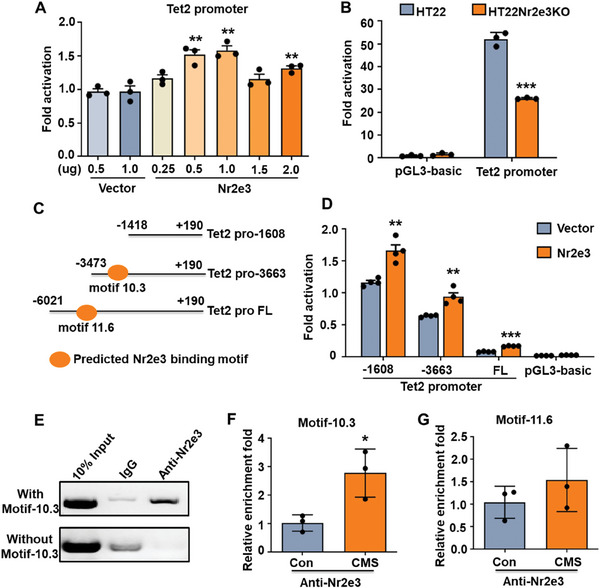
Nr2e3 bound to the Tet2 promoter directly and enhanced the activity of the Tet2 promoter. A) Different doses (0.25, 0.5, 1.0, 1.5, and 2.0 µg) of Nr2e3 plasmid were co‐transfected with full‐length Tet2 promoter (Tet2 pro FL) in 293T cells. Dual‐luciferase reporter assay was performed to measure the activity of the Tet2 promoter at 48 h after transfection (n = 3). B) Full‐length Tet2 promoter plasmid was transfected into HT22 Nr2e3KO cell line. Dual‐luciferase reporter assay was performed at 48 h after transfection (n = 3). C) The predicted Nr2e3 binding motifs (motif 10.3 and motif 11.6) at the Tet2 promoter by Jaspar (vs UCSD NCBI37 mm9) were shown. Different Tet2 promoter plasmids containing different truncated Tet2 promoters were constructed. D) Truncated Tet2 promoter plasmid and control plasmids were transfected into 293T cells. Dual‐luciferase reporter assay was performed to measure the activity of Tet2 promoter at 48 h post‐transfection (n = 3). E) Full‐length Tet2 promoter plasmid (with motif‐10.3) and Tet2 promoter plasmid without motif‐10.3 (Tet2 pro‐1608) were separately co‐transfected into HEK293T cells with Nr2e3 plasmid. Nr2e3 monoclonal antibody and IgG antibody were used to perform the CHIP. The captured genomic DNA by Nr2e3 antibody was used to amplify PCR product containing Tet2 motif‐10.3. F, G) The hippocampus of CMS mice and control mice were collected. Nr2e3 monoclonal antibody was used to perform the CHIP‐qPCR in the hippocampal lysates. The enrichment fold changes of F) motif‐10.3 and G) motif‐11.6 were analyzed by qPCR (n = 3). All data were presented as mean ± SEM. A) by using one‐way ANOVA with Tukey's multiple comparison tests. B, D) by using two‐way ANOVA with Tukey's multiple comparison test. F, G by using Student's t‐test (*p < 0.05, **p < 0.01, and ***p < 0.001).
