Figure 1.
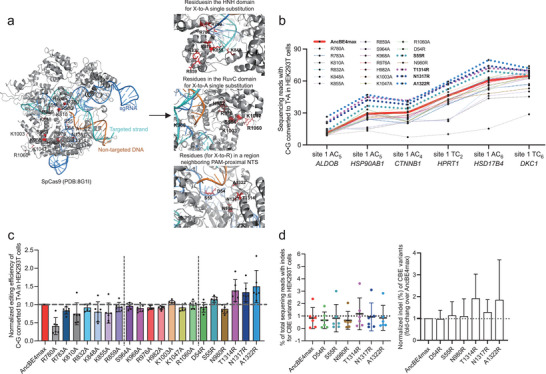
Structure‐guided engineering of nCas9 to increase AncBE4max editing efficiencies. a) On the left, the positions of individual point mutations tested are shown on the solved structure of SpCas9 in complex with sgRNA and target DNA (PDB: 8G1I). The locations of the focused amino acids are highlighted in red color with black letter labels. On the right, the zoomed‐in structures show part of the HNH domain, RuvC domain, and a region near the initially unwound non‐targeted strand. The latter contains the amino acid residues where individual X‐to‐R substitutions were made. b) Parallel comparison of editing activities by AncBE4max and 20 single‐substitution, engineered nCas9/AncBE4max variants at six genomic loci in HEK293T cells. The results show the percentage of C‐to‐T conversion at the mainly edited position within the editing window. The AncBE4max and four single substitution groups (S55R, T1314R, N1317R, and A1322R) with increased editing efficiencies are highlighted in bold. Data are presented as means ± s.d. (n = 3 biological replications). c) Results in (b) were further analyzed by considering editing efficiency at all sites (means ± s.d., n = 6 sites) as a whole. The editing levels induced by the control AncBE4max at each site were set as 1 (position marked by horizontal gray dashed line). d) The percentages of editing‐associated indels at six target sites by the six X‐to‐R variants are shown (see legend of (a, b) for reference). The left graph corresponds to the true indel percentages (means ± s.d.). The indel levels normalized to those by AncBE4max are shown on the right (error bar: s.d.).
