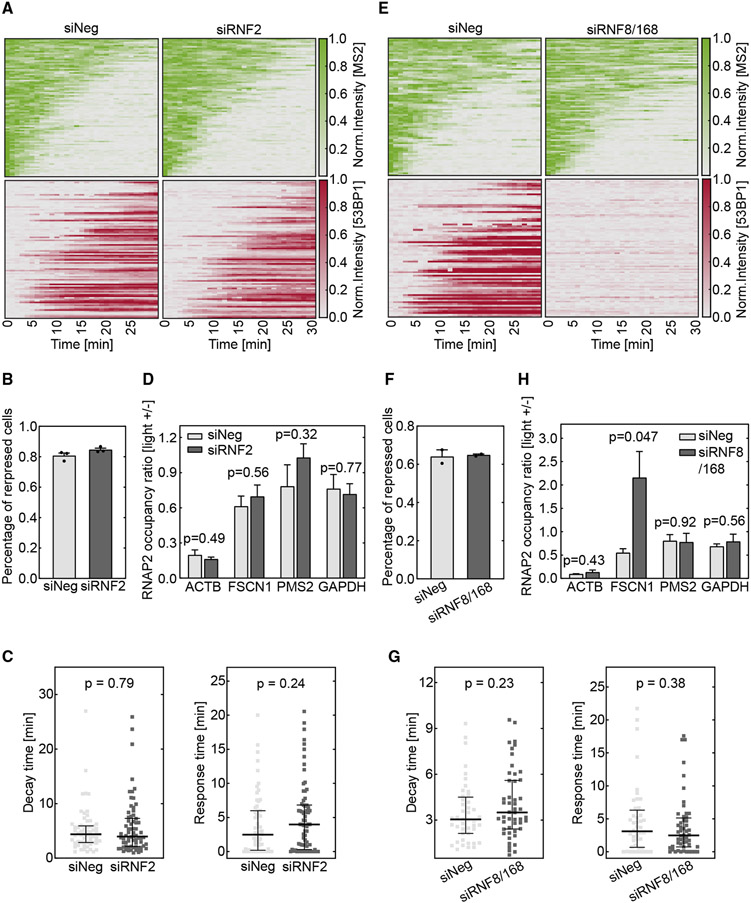
Figure 3. DSB-induced transcription repression does not depend on PRC1- or RNF8/168-mediated ubiquitination.
Cells were treated with siRNAs to knock down RNF2 (A–D) or RNF8/168 (E–H). The DSB was induced by vfCRISPR in the live-cell transcription reporter (Figure 1A) stably integrated in U-2 OS cells (A–C and E–G) or endogenous ACTB in HEK293T cells (D and H).
(A and E) Heatmaps of TS and 53BP1 intensities were measured from live-cell imaging in the same way as in Figure 1. (A) RNF2 knockdown samples (siRNF2, n = 89 cells) and negative control (siNeg, n = 91 cells). Cells with recruited 53BP1 were selected. (E) RNF8 and RNF168 knockdown samples (siRNF8/168, n = 94 cells) and negative control (siNeg, n = 80 cells). All stimulated cells were analyzed since there was no 53BP1 recruitment.
(B and F) Percentages of repressed cells were measured for knockdown samples and control samples during 30 min tracking. Cells were deemed repressed if the averaged TS intensities in the last 4 frames were lower than a threshold value determined from siNeg control cells with 53BP1 recruitment (STAR Methods). (B) RNF2 knockdown samples and negative control (n = 3 biological replicates). (F) RNF8/168 knockdown samples and negative control (n = 2 biological replicates). Error bar: SEM.
(C and G) Transcription decay and response times were obtained by fitting the delayed decay model (Figure 1G) to the repressed TS intensities in (B) and (F). (C) RNF2 knockdown samples (n = 70 cells) and negative control (n = 62 cells). (G) RNF8/168 knockdown samples (n = 48 cells) and negative control (n = 44 cells). Error bar: quartiles. Unpaired Mann-Whitney test was performed.
(D and H) ChIP-qPCR experiments of RNAPII were performed in HEK293T cells when the ACTB locus was damaged by vfCRISPR. Change of RNAPII occupancy (measured in reads per million [RPM]) upon light stimulation on the ACTB locus and nearby genes was measured. GAPDH on a different chromosome served as a control gene to indicate global transcriptome change upon UV exposure. (D) RNF2 knockdown samples and negative control (n = 3 biological replicates). (H) RNF8/168 knockdown samples and negative control (n = 3 biological replicates). FSCN1: 70 kb from DSB. PMS2: 487 kb from DSB. Error bar: SEM. Unpaired t test was performed.