Figure 6.
Axonal ER-ribosome interactions influence axonal calreticulin translation, and P180 interacts with specific mRNAs in neurons
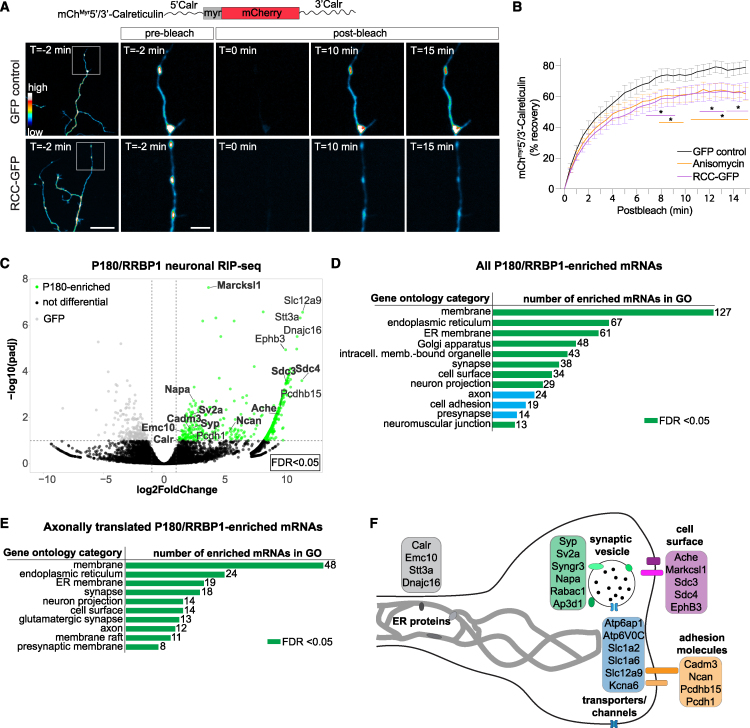
(A) Schematic diagram of the calreticulin FRAP reporter construct and representative pre- and post-bleach images of the calreticulin FRAP reporter in distal axons of DIV4 and -5 neurons. Scale bars represent 20 μm (left column) and 5 μm (right columns).
(B) Quantification of FRAP assays in distal axons of mChMyr5′/3′-Calreticulin (average % recovery) in GFP, RCC-GFP, and anisomycin-treated conditions.
(C) Volcano plot showing differential gene expression analysis of mRNAs pulled down with P180, compared with GFP control. Gene names are indicated, and bolded names indicate that they have been previously detected in axonal translatome studies.4,27
(D) GO analysis of P180-enriched mRNAs identified by differential gene expression analysis. The number of genes in each category is plotted and noted after each bar. Green bars represent significantly enriched GO categories.
(E) GO analysis of P180-enriched mRNAs that are also known to be axonally translated.4,27 The number of genes in each category is plotted and noted after each bar. Green bars represent significantly enriched GO categories.
(F) Schematic representation of an axon/growth cone/pre-synapse highlighting known axonally translated mRNAs and their functional categories of mRNAs enriched after P180 pull-down.
Line graph in (B) represents the mean ± SEM of recovery of 18 (GFP), 10 (RCC-GFP), or 12 (anisomycin) neurons per condition from two independent experiments. ∗p < 0.05 comparing conditions to each other using a 2-way ANOVA test.