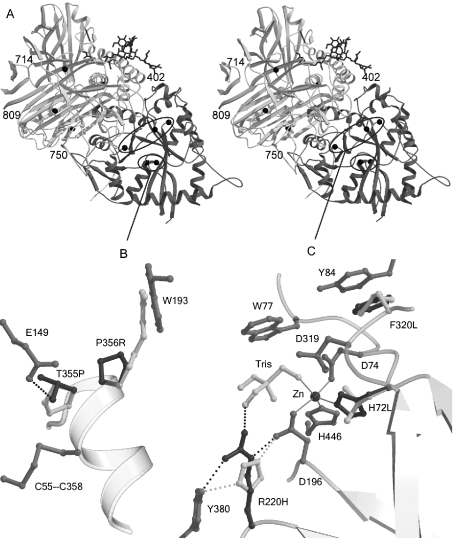
Figure 6. Amino acid substitutions modelled in the bovine LAMAN structure.
(A) Stereographic representation of the complete structure. The active-site domain is darker than the rest of the structure. The affected residues are indicated with black balls. Mutations outside the active-site domain are labelled, and mutations in the N-terminal domain are shown in detail in (B, C). (B) Mutations affecting the folding of LAMAN. The mutated wild-type residues are shown in dark grey and the modelled mutations in white. Hydrogen bonds are displayed with broken lines and metal co-ordination with solid lines. The T355P mutation disrupts a hydrogen bond to Glu149 and, probably, to the free cysteine residue Cys358, which in the mature enzyme participates in a disulphide bridge. Pro356 is in optimal position to initialize helix formation and is important for the rate of folding of LAMAN. (C) Mutations that inactivate the enzyme, but allow folding, are located close to the active site: although His220 hydrogen-bonds to the nucleophile Asp196, it cannot co-ordinate the substrate as Arg220 probably does. His72 is involved in the metal binding, whereas Phe320 follows the active-site residue Asp319 and makes a stacking interaction with Tyr84. Probably, the hydrophobic stacking also stabilizes Trp77, which is involved in substrate binding.