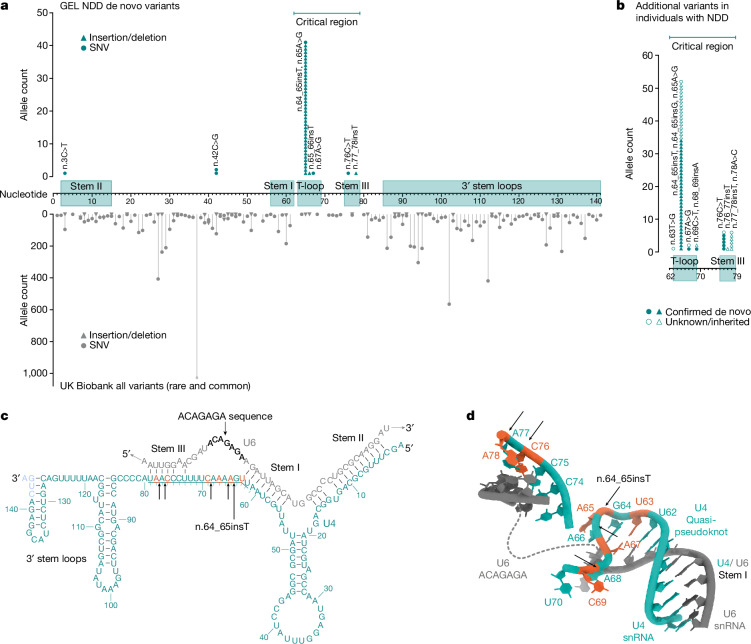
Fig. 1. A highly structured 18 bp region of RNU4-2 that is critical for BRR2 helicase activity is enriched for variants in NDD and depleted in population cohorts.
a, Allele counts of de novo variants in 8,841 undiagnosed NDD probands in GEL (top; teal) and the UK Biobank cohort (bottom; grey) across RNU4-2. The 18 bp critical region, which is depleted of variants in the UK Biobank, is marked by a horizontal bar at the top of the plot. b, Allele counts of further variants identified in individuals with NDD in the critical 18 bp region. This includes 16 individuals with seven variants without sequencing data for both parents in GEL and variants identified in individuals from the following extra cohorts (Methods): NHS GMS (n = 19); MSSNG (n = 2); SSC (n = 1); GREGoR (n = 10) and UDN (n = 6); from personal communication or Matchmaker Exchange (n = 16). c, Schematic of U4 (teal) binding to U6 snRNA (grey). The 18 bp critical region is underlined. Nucleotides 142 to 145 of U4 (in blue) are not within the GENCODE transcript of RNU4-2 but are included in previous figures of the U4/U6 duplex in the literature on which this depiction is based38 and are present in the RNA-seq reads from human prefrontal cortex in BrainVar. d, The structure of U4 and U6 snRNAs resolved by cryogenic electron microscopy18. U4 residues in the critical region are labelled with the reference nucleotide and numbered according to the position along the RNA (for example, U62 indicates a uracil residue in the reference sequence at position 62). Created using publicly accessible coordinates from the RCSB Protein Data Bank39 (PDB structure 6QW6). In both c and d, single base insertions identified in individuals with NDD are shown by black arrows and positions of SNVs by orange nucleotides.