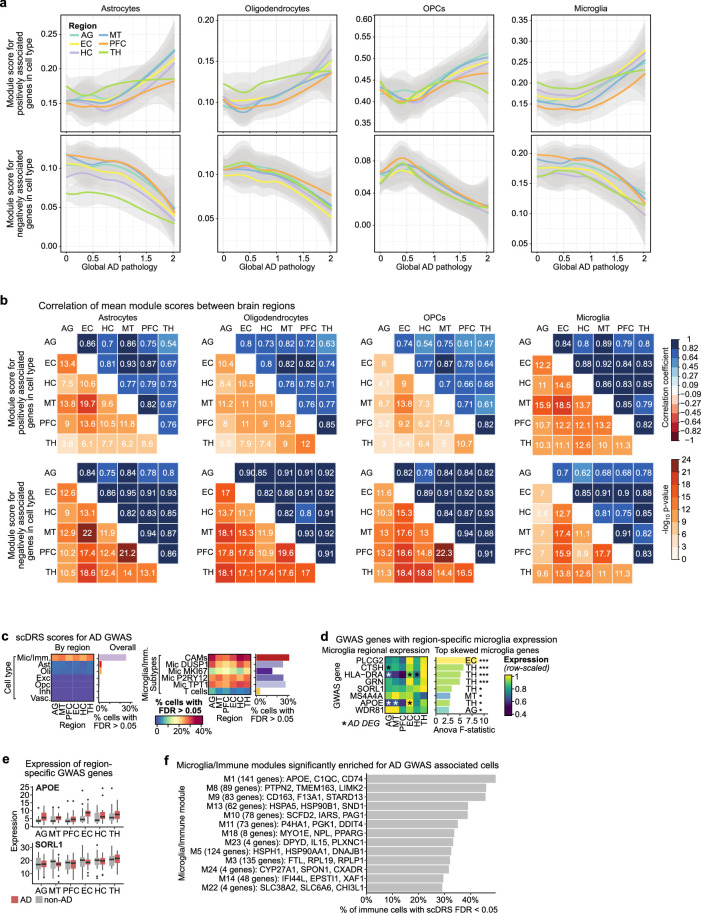
Extended Data Fig. 9. Inter-regional comparison of AD pathology-associated gene sets and region-specific GWAS enrichments.
a-b, Seurat module scores of genes significantly positively (top) or negatively (bottom) associated with the global AD pathology variable in prefrontal cortex for astrocytes, oligodendrocytes, OPCs, and microglia across brain regions and the spectrum of global AD pathology burden. The gene sets used for computing the module scores (genes significantly associated with global AD pathology burden) were determined based on snRNA-seq data derived from prefrontal cortex tissue of 427 ROSMAP study participants. The scatterplots (a panels) illustrate the relationship between global AD pathology burden and the mean module score for the specified gene set, with this mean score calculated by averaging the module scores of all cells of the designated cell type isolated from an individual. A LOESS (Locally Estimated Scatterplot Smoothing) regression line with a 95% confidence interval is shown, and the regression lines are coloured by brain region. The central LOESS regression line represents the local measure of central tendency, calculated through locally weighted regression to reflect the smoothed relationship between the module scores indicated and global AD pathology burden. Interregional Pearson’s correlation analysis of mean module scores (b panels) was performed by first averaging the module scores of all cells of the cell type of interest from an individual study participant. The correlation analysis was then performed between regions based on these averaged scores. P values were calculated using the cor.test function in R and were adjusted for multiple testing using the p.adjust function with the Benjamini-Hochberg method. c, Heatmap (by region, left) and barplot (over all regions, right) showing the percentage of cells with significant scDRS (disease relevance scores) for AD GWAS. Rows are split into major cell type groups (top) and microglia and immune subtypes. d, Regional expression (heatmap, left) and F-statistic for region in predicting expression (barplot, right) for eight GWAS genes with significantly region-specific expression in microglia. Barplot is coloured by the top expressed region (regression coefficient). Heatmap is labelled with stars if the gene is a DEG for that region. e, Boxplots showing expression of two of the region-specific GWAS genes in individuals with and without a pathologic diagnosis of AD. f, Microglia/immune modules associated with AD GWAS. Fraction of microglia or immune cells with significant expression of each module (z-score > 2.5) and with significant scDRS scores (FDR < 0.05). Only significant modules are shown (adjusted p < 0.01, hypergeometric test with BH correction).