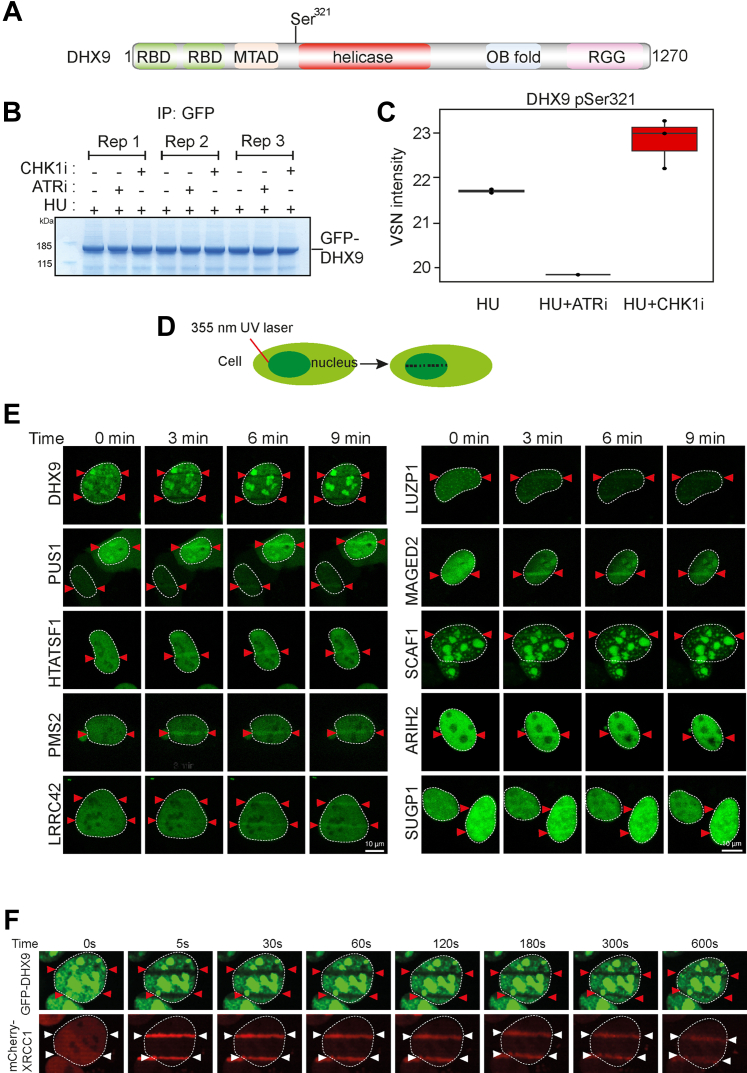
Fig. 3.
Screening novel ATR targets for recruitment to DNA damage sites. A, schematic diagram showing the domain organization of DHX9. RBD, RNA binding domain; MTAD, minimal transcriptional activation domain; RGG, RGG-rich domain; OB fold, oligonucleotide/oligosaccharide binding fold. B, U-2 OS cells were transfected with GFP-tagged DHX9 and after 24 h cells were lysed, and cell extracts were subjected to immunoprecipitation with anti-GFP-agarose beads. Precipitates were subjected to SDS-PAGE and staining with Coomassie Brilliant Blue; the bands corresponding to the GFP-DHX9 were excised and processed for mass spectrometric detection of relevant phospho-peptides. Three independent co-transfection experiments were done for every condition (Rep = biological replicate). C, label-free quantification was used to generate a boxplot showing VSN transformed intensity of phospho-peptides containing to DHX9 pSer321. Mass spectrometry raw data was uploaded to ProteomeXchange via the PRIDE partner repository (126) and can be downloaded via the identifier PXD041250. Data analysis scripts and annotated spectra (66) can be accessed via Zenodo under https://doi.org/10.5281/zenodo.10882997. D, schematic diagram showing micro-irradiation of BrdU-sensitized cells to induce DNA damage along a track in the nucleus. E, BrdU–sensitized U-2 OS cells transiently expressing GFP-tagged forms of the proteins indicated were line micro-irradiated and imaged after 2 min. F, BrdU-sensitized U-2 OS cells stably expressing mCherry-XRCC1 and expressing GFP-DHX9 in a tetracycline-inducible manner were subjected to laser micro-irradiation and live imaged at the times indicated.