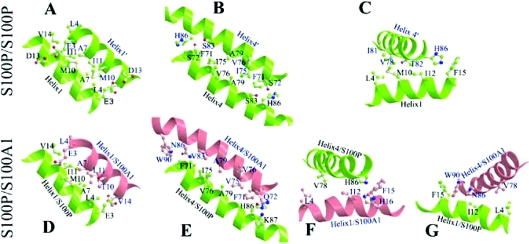
Figure 7. Comparison of the S100P homodimeric interface and modelled heterodimeric interface with S100A1.
Ball-and-stick ribbon models of interacting helices were generated by MOLSCRIPT software for the interaction between self-associated S100P (A–C) and for dimerization with S100A1 (D–G) modelled using CHARMM software, as described in the Experimental section. Interacting amino acid residues are shown as ball and stick molecular representations. Interactions are shown between helices 1 of the interacting partners (A, D), between helices 4 of the interacting partners (B, E), and between helix 4 and helix 1 of S100P for the self-association (C), and between helix 4 of S100P and helix 1 of S100A1 (F) and between helix 1 of S100P and helix 4 of S100A1 (G) for the heterodimeric interaction. The indicated amino acids are supposed to contribute to the stability of the dimer interface.