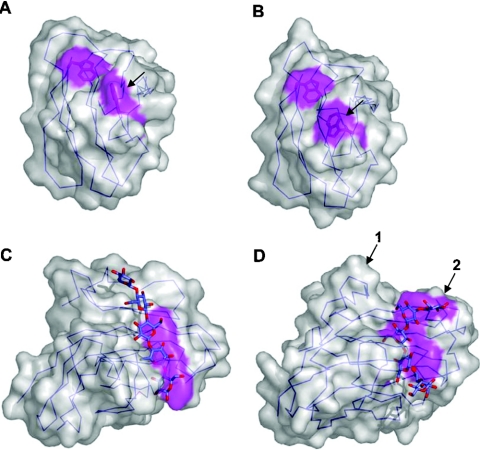
Figure 5. Binding-site topography and oligosaccharide recognition.
Surface representations of the NMR structures of wild-type CfCBM2b-1 (A) and the same protein with an Arg262→Gly mutation (B). The arrow indicates the tryptophan residue that changes conformation due to the mutation. The binding site of CfCBM4-1 extend in a straight path across the face of one of the β-sheets of this CBM creating a linear binding site appropriate for binding cello-oligosaccharides (C). In TmCBM4-2, which has significant sequence identity with CfCBM4-1, two loops are extended (D); one to block an end of the binding site (shown by the arrow labelled 1) and another to accommodate the curvature of the laminarioligosaccharide (shown by the arrow labelled 2). Solvent-accessible surfaces are shown in transparent grey with surfaces created by the aromatic amino acid side chains involved in binding shown in magenta. C-α traces are shown in blue.