Figure 3.
Proteomic analysis using co-immunoprecipitation reveals reduced interactions between HDAC3 variants and the subunits of NCoR and CoREST complexes
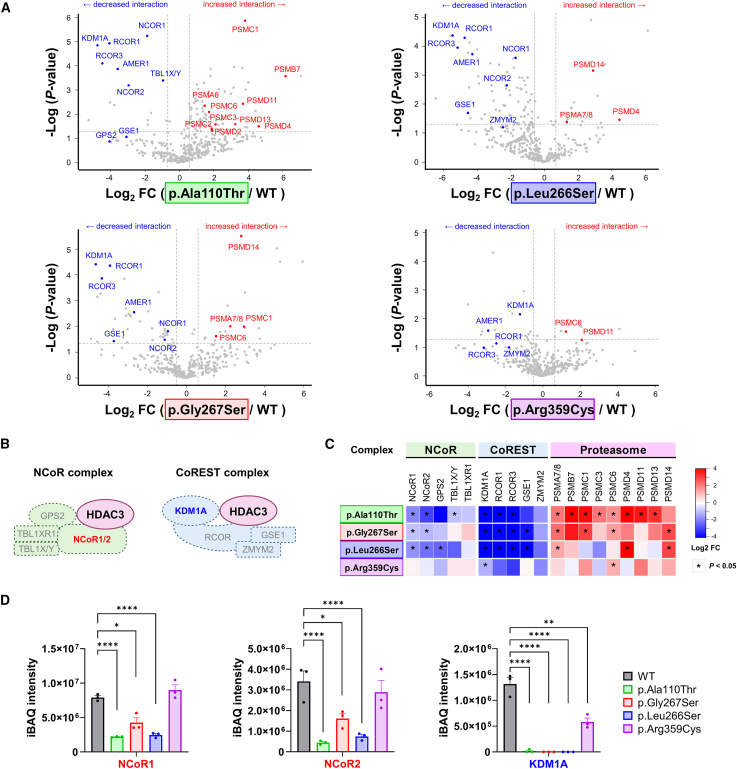
(A) Volcano plots of protein interaction profiles for four selected HDAC3 variants (p.Ala110Thr, p.Gly267Ser, p.Leu266Ser, and p.Arg359Cys) compared to HDAC3 wild-type (WT) in HEK293T cells. The plots highlight important proteins involved in the NCoR and CoREST complexes or the proteasome pathway, with the x axis representing the log2 fold change (FC) and the y axis showing the −log10p value (measured in triplicate for each data point).
(B) Schematic presentation of the NCoR and CoREST complexes, with the position of HDAC3 and its interacting subunits within each complex.
(C) A heatmap summarizes the log2 FC in protein interactions for HDAC3 variants relative to WT. Each column represents a distinct protein, as highlighted in (A), and each row corresponds to the four tested HDAC3 variants. Blue shades denote decreased, and red shades indicate increased interaction strength. The data show decreased interactions with subunits of the NCoR and CoREST complexes and contrasted with an increased interaction tendency with subunits of the proteasome pathway, illustrating variant-specific effects on HDAC3 function and complex integrity.
(D) Bar graphs display quantified protein levels using the intensity-based absolute quantification (iBAQ) method for three major proteins: NCoR1, NCoR2, and KDM1A. The data reveal a consistent reduction in association with the HDAC3 variants for these proteins. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001. Data are plotted as mean ± SD (n = 3/data point).