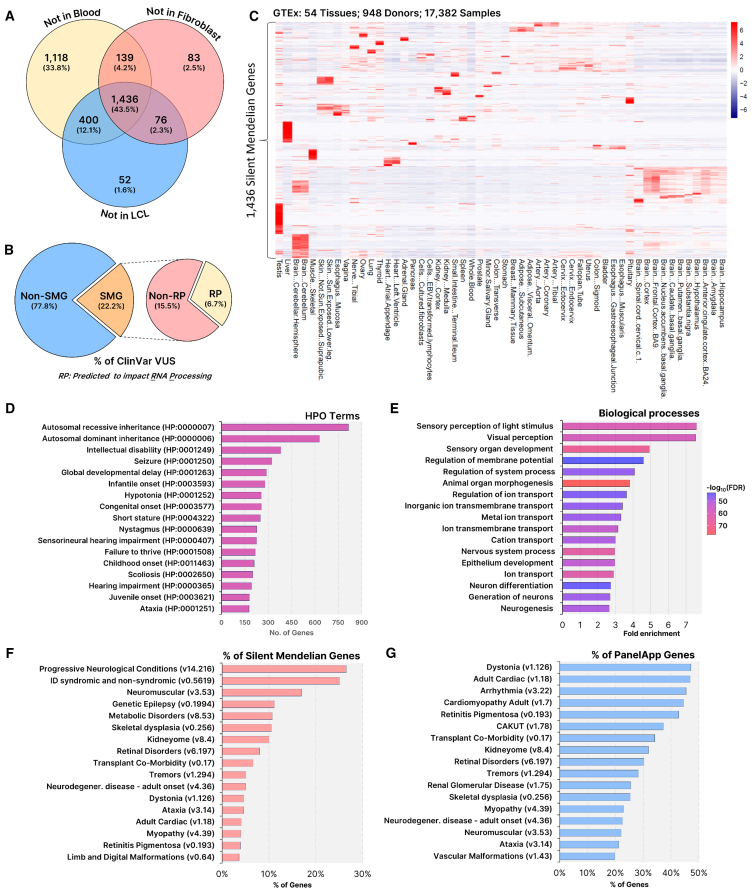
Figure 1.
Silent Mendelian genes have restricted tissue expression and are prominently involved in neurological disorders
(A) 1,436 Mendelian genes are silent. Analysis of 4,878 Mendelian disease genes (Nijmegen DG Panel 3.2.0) using minimum required sequencing depth (MRSD) identified 1,436 genes that are not sufficiently expressed in whole blood, LCLs, or HDFs for the purpose of conducting robust analysis of mRNA splicing using srRNA-seq. These genes are termed silent Mendelian genes (SMGs).
(B) Large numbers of VUS are found in SMGs. From the catalog of VUSs in ClinVar, 22.2% of all are found in SMGs, of which ∼30% are predicted to impact RNA processing (RP; 6.66% of all ClinVar VUSs).
(C) SMGs display highly restricted tissue-specific expression. Heatmap showing the level of mRNA expression (TPM) of each of the 1,436 SMGs across 54 different tissues taken from 948 donors (data obtained from GTEx Version 8).
(D–G) Phenotypes, disease categories, and biological processes associated with SMGs. (D) Most frequently associated human phenotype ontology (HPO) terms. (E) Top-ranked gene ontology (GO) biological processes (analyzed via ShinyGO 0.77 using whole-genome background, ranked by Fold enrichment and false discovery rate [FDR]).
(F–G) Disease types ranked based on (F) their contribution to the number of SMGs or (G) on the proportion of known associated genes that are silent. Disease gene lists referenced from PanelApp Australia (accessed December 12, 2023).