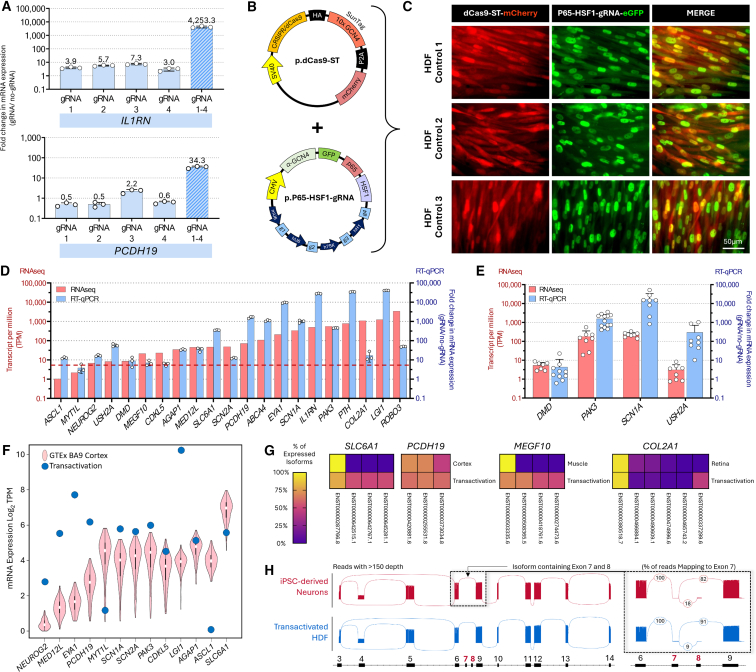
Figure 3.
Robust Transactivation of SMGs in HDFs
(A) Comparison of transactivation levels of IL1RN and PCDH19 using single gRNAs versus a multiplex of four gRNAs. Relative gene expression analyzed via real-time qPCR with values normalized to ACTB and expressed relative to the negative control (dCas9-ST-PH-no gRNA). Error bars respresent standard deviation.
(B) The dCas9-ST-PH-gRNA complex was engineered across two lentiviral transgenes with fluorescent reporters.
(C) Optimized transient delivery of dCas9-ST-PH-gRNA complex to HDFs. Highly efficient lentiviral co-delivery of dCas9-ST and P65-HSF1-gRNA transgenes in three control HDFs. Representative images showing co-expression of p.dCas9-ST transgene (mCherry) and p.p65-HSF-gRNA (eGFP) 72 h after transduction.
(D) Co-expression of dCas9-ST-PH complex and 4 gRNAs successfully transactivates expression of many SMGs in HDFs. Bar graph showing the individual transactivation levels of 20 SMGs and IL1RN mediated by co-expression of dCas9-ST-PH complex and four gRNAs. Expression levels (TPM) generated from srRNA-seq (red) and relative gene expression generated from real-time qPCR (blue) with values normalized to ACTB and expressed relative to negative control (dCas9-ST-PH with no gRNA). Error bars respresent standard deviation. Red dotted line corresponds to TPM = 5.
(E) SMGs can be robustly activated across multiple experiments and HDFs. Bar graph showing the transactivation of DMD, PAK3, SCN1A, and USH2A mediated by dCas9-ST-PH-gRNA in multiple different HDF lines. The bar graph data presents the mean and standard deviation from the biological replicates, with each dot plot representing a different cell line. Data presented are expression levels (TPM) generated from srRNA-seq (red bars) and relative gene expression generated from real-time qPCR with values normalized to ACTB and expressed relative to the negative control (dCas9-ST-PH with no gRNA; blue bars).
(F) Transactivated SMGs expression levels are comparable to endogenous expression levels in CRTs. Violin plots show endogenous expression of a subset of SNGs in the adult cerebral cortex. The blue dots show the expression of the same genes transactivated in HDFs. Data presented are expression levels (TPM) calculated independently for cortex data accessed from GTEx (Version 8) and transactivated HDF data generated from srRNA-seq, respectively.
(G) Transactivated genes in HDFs express diverse isoforms. Comparison isoforms expressed in CRTs (extracted from GTEx Version 8) with transactivated HDFs (srRNA-seq).
(H) Complex and rare splicing events are observed using transactivation. Sashimi plot displaying complex splicing patterns of PAK3 in iPSC-derived neurons recapitulated following transactivation in HDFs. Only events with read depth greater than 150 are shown. Insert highlights rare isoform containing exon 6 and 7. All reads that map to exon 6 are shown.