Figure 4.
Investigation of RNA variants in SMGs using transactivation of HDFs
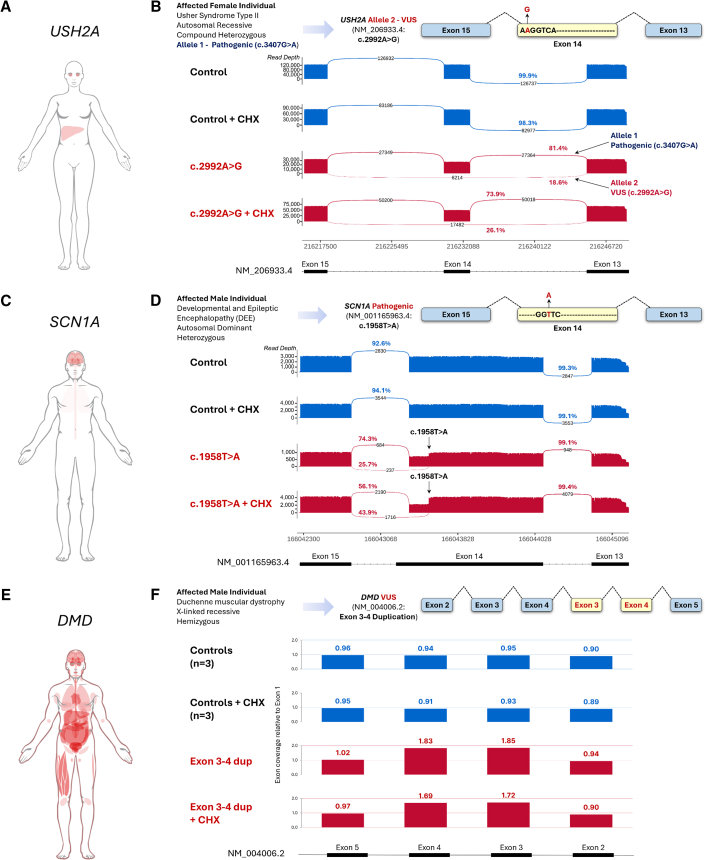
(A) Illustration of USH2A mRNA expression (red) in human adult tissues as reported by the Human Protein Atlas (HPA).
(B) Diagram depicts the USH2A variant under investigation. Sashimi plots report USH2A mRNA splicing. Data derived from Oxford Nanopore long read sequencing of RT-PCR amplicons (exons 13–16) produced using RNA isolated following transactivation of USH2A in HDFs derived from healthy control and affected individuals in the presence and absence of cycloheximide (CHX). Arrows in the sashimi plots specify the reads coming from the alleles with pathogenic variant and allele with VUS as segregated by allelic phasing.
(C) Illustration of SCN1A mRNA expression (red) in human adult tissues (HPA).
(D) Diagram depicts the SCN1A variant under investigation. Sashimi plots report SCN1A mRNA splicing. Data derived from Oxford Nanopore long-read sequencing RT-PCR amplicons (exons 13–17) produced from RNA isolated following transactivation of SCN1A in HDFs derived from healthy control and affected individuals in the presence and absence of CHX. Arrows on the sashimi plot indicates the position of the pathogenic variant.
(E) Illustration of DMD mRNA expression (red) in human adult tissues (HPA).
(F) Diagram depicts the DMD variant under investigation. Graphs represent relative read depth of reported across DMD exons 2–5 as determined using long read sequencing of RT-PCR amplicons produced from RNA isolated following transactivation of DMD in HDFs derived from healthy control and affected individuals in the presence and absence of CHX. Note read depth is 1.8 times greater (∼double) in exons 3– and 4 only in samples from the affected individual and is not influenced by CHX.