Figure 5.
Transdifferentiation of HDFs directly into iNeurons
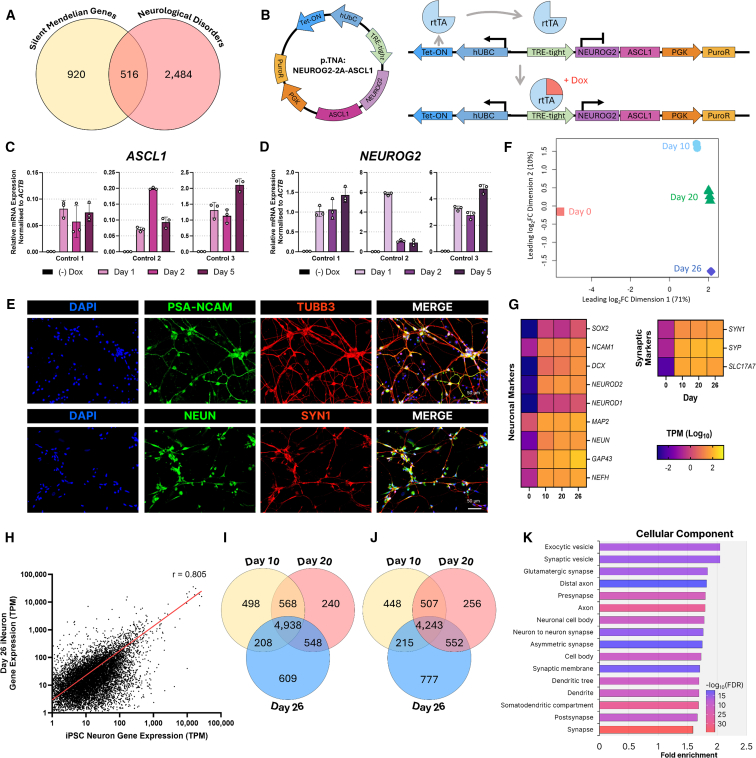
(A) 516 neurological disorder genes are silent. A comparison between silent mendelian genes (SMGs) and a list of 3,000 neurological disorders reveals an overlap of 516 genes. These genes, herein referred to as silent neurological genes (SNGs), are not expressed at sufficient levels in CATs of blood, LCLs, or HDFs to enable analysis of mRNA splicing using srRNA-seq.
(B) Schematic of the vector transgene featuring a Tet-On inducible promoter driving overexpression of NEUROG2 and ASCL1 (abbreviated as TNA). In the TNA transgene, the human ubiquitin C (hUbC) promoter drives the expression of Tet-ON encoding the reverse tetracycline-controlled transactivator (rtTA). rtTA binds the TRE-tight promoter when in the presence of doxycycline thus inducing NGN2 and ASCL1 expression. The phosphoglycerate kinase promoter (PGK) drives the constitutive expression of a puromycin-resistance cassette. The TNA transgene can be packaged into lentivirus.
(C and D) Fibroblasts transduced with TNA overexpress NERUOG2 and ASCL1 in response to doxycycline treatment. RT-qPCR performed on RNA isolated from 3 control HDF lines transduced with TNA and treated with or without 2 μg/mL doxycycline (dox) for 1, 2, and 5 days (C) ASCL1 expression and (D) NEUROG2 expression. Error bars respresent standard deviation.
(E) iNeurons display overt neuronal morphology and express a set of neuronal marker proteins. Immunofluorescent imaging of day 22 control iNeurons: PSA-NCAM (green), TUBB3 (red), NeuN (green), SYN1 (red), DAPI (blue). Scale bars, 50 μm.
(F) Principal component analysis (PCA) of srRNA-seq. RNA was collected at day 0, 10, 20, and 26 of transdifferentiation. Experiment done in quadruplicate. Note that ∼70% of the transcriptional variance occurs by day 10 of transdifferentiation.
(G) The srRNA-seq analysis reveals that iNeurons express cohorts of neuronal cell and synapse marker genes. Expression is reported as TPM (log10).
(H) The transcriptional profile of iNeurons correlates with iPSC-derived neurons. The expression of genes (>1 TPM, n = 11,119 genes) was correlated between iNeurons (day 26, n = 4) and iPSC-derived neurons (day 90 of iPSC neuronal differentiation, n = 1) using Pearson’s correlation (r = 0.805, p < 0.0001).
(I and J) Differential gene expression analysis of iNeuron transdifferentiation. The srRNA-seq transdifferentiation data were used to identify differentially expressed genes between HDFs (day 0) and other time points (day 10, 20, and 26) during transdifferentiation and the overlapping genes of each comparison identified. (I) Comparison of upregulated genes. (J) Comparison of downregulated genes.
(K) Gene ontology
analysis of genes upregulated in iNeurons at day 26 of transdifferentiation performed using ShinyGO 0.77. The highest-ranking GO terms are reported as fold enrichment and the FDR (-log10FDR).