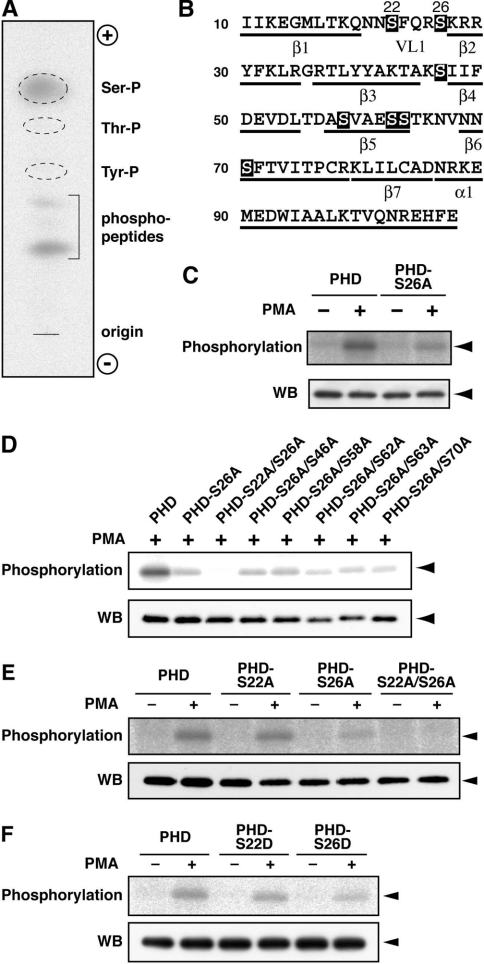
Figure 2. Phosphorylation at Ser-22 and Ser-26 in the DGKδ1-PH domain.
(A) Phosphoamino acid analysis of the DGKδ1-PH domain. The analysis was performed as described in the Experimental section. Ser-P, phosphoserine; Thr-P, phosphothreonine; Tyr-P, phosphotyrosine; phosphopeptides, undigested phosphopeptides. (B) Primary and predicted secondary structure of the DGKδ1-PH domain. Serine residues are highlighted. α, α-helix; β, β-sheet; VL1, variable loop 1. (C–F) Effects of point mutations on phosphorylation of the DGKδ1-PH domain. COS-7 cells were transfected with expression plasmids as indicated. After starvation and 32P-labelling, cells were incubated for 30 min in the presence of 100 nM PMA (+) or 0.1% DMSO (−). FLAG-tagged proteins were immunoprecipitated with anti-FLAG antibody and analysed by SDS/PAGE. The radioactive signal was visualized by a BAS1800 Bio-Image Analyzer (upper panels), and the immunoprecipitated 3×FLAG-tagged proteins were analysed by Western blotting using anti-FLAG antibody (lower panels). The position of the DGKδ1-PH domain and its mutants is indicated by arrowheads. Representatives of three repeated experiments are shown. PHD, PH domain.