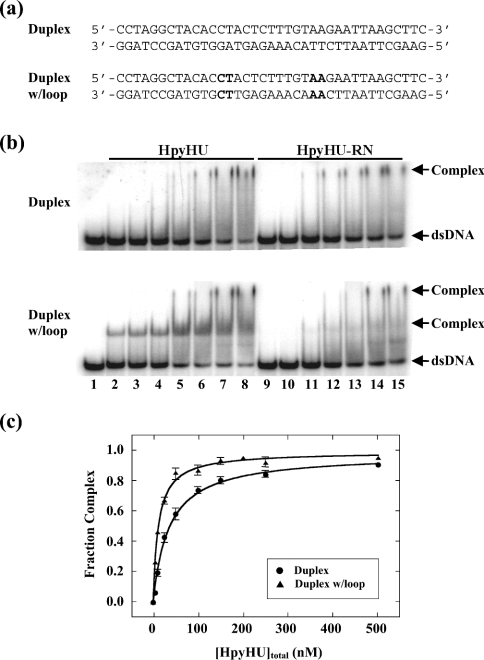
Figure 8. EMSA of HpyHU binding to 37 bp DNA.
(a) Sequence of the DNA probes. The sequence corresponds to the thymine-containing version of a preferred binding site for the B. subtilis bacteriophage SPO1-encoded HU homologue TF1. Loops are generated by substituting the sequence of the bottom strand to generate tandem mismatches of identical opposing bases. Sequences generating loops are in bold. (b) Titrations of HpyHU (left panel) and HpyHU-RN (right panel) with perfect duplex DNA (upper panel) and loop-containing DNA (lower panel). Complex and free DNA is identified at the right. HpyHU variants are identified at the top, and protein concentrations (identical for both panels) are as follows: Lane 1, no protein; lanes 2–8 and 9–15, reactions with 5, 10, 25, 50, 100, 250, 500 nM protein respectively (from left to right). (c) Binding isotherm for HpyHU binding to 37 bp duplex DNA (•) or duplex with loops (▴), corrected for complex dissociation during electrophoresis.