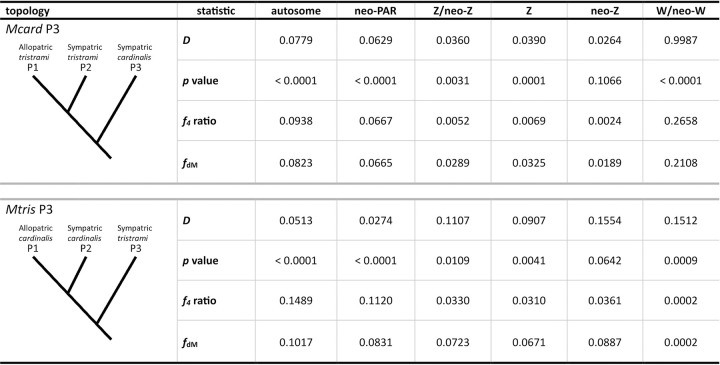
Fig 4. Test of introgression, ABBA-BABA results.
A positive D statistic indicates gene flow between P2 and P3, and the p value specifies whether D is significantly different from zero, using block-jackknife procedure. The f4 ratio is calculated by splitting P3 population into two subsets to calculate the admixture proportion, assuming unidirectional introgression P3 → P2. The average fdM statistic is calculated using 100 SNP non-overlapping windows (W/neo-W calculated in 50 SNP non-overlapping windows) where fdM ≥ 0, indicating either no gene flow (fdM = 0) or gene flow between P2 and P3 (fdM > 0). Allopatric M. cardinalis includes samples from both Ugi and Three Sisters. M. pulchella is the P4 outgroup in both topologies.