Abstract
The Bacillus licheniformis 749/I BlaI repressor is a prokaryotic regulator that, in the absence of a β-lactam antibiotic, prevents the transcription of the blaP gene, which encodes the BlaP β-lactamase. The BlaI repressor is composed of two structural domains. The 82-residue NTD (N-terminal domain) is a DNA-binding domain, and the CTD (C-terminal domain) containing the next 46 residues is a dimerization domain. Recent studies have shown the existence of the monomeric, dimeric and tetrameric forms of BlaI in solution. In the present study, we analyse the equilibrium unfolding of BlaI in the presence of GdmCl (guanidinium chloride) using different techniques: intrinsic and ANS (8-anilinonaphthalene-l-sulphonic acid) fluorescence, far- and near-UV CD spectroscopy, cross-linking, analytical ultracentrifugation, size exclusion chromatography and NMR spectroscopy. In addition, the intact NTD and CTD were purified after proteolysis of BlaI by papain, and their unfolding by GdmCl was also studied. GdmCl-induced equilibrium unfolding was shown to be fully reversible for BlaI and for the two isolated fragments. The results demonstrate that the NTD and CTD of BlaI fold/unfold independently in a four-step process, with no significant co-operative interactions between them. During the first step, the unfolding of the BlaI CTD occurs, followed in the second step by the formation of an ‘ANS-bound’ intermediate state. Cross-linking and analytical ultracentrifugation experiments suggest that the dissociation of the dimer into two partially unfolded monomers takes place in the third step. Finally, the unfolding of the BlaI NTD occurs at a GdmCl concentration of approx. 4 M. In summary, it is shown that the BlaI CTD is structured, more flexible and less stable than the NTD upon GdmCl denaturation. These results contribute to the characterization of the BlaI dimerization domain (i.e. CTD) involved in the induction process.
Keywords: BlaI Bacillus licheniformis, domain structure, folding intermediate, guanidinium chloride, β-lactamase induction, protein folding
Abbreviations: ANS, 8-anilino-1-naphtalene sulphonic acid; AU, analytical ultracentrifugation; CTD, C-terminal domain; DSP, dithiobis(succinimidyl propionate); GdmCl, guanidinium chloride; HSQC, heteronuclear single-quantum coherence; MALDI–TOF MS, matrix-assisted laser-desorption ionization–time-of-flight MS; Ni-NTA, Ni2+-nitrilotriacetate; NTD, N-terminal domain; Rs, Stokes radius; SEC, size-exclusion chromatography; 3D, threedimensional; Ve, elution volume
INTRODUCTION
The Bacillus licheniformis 749/I BlaI repressor is a prokaryotic regulator that, in the absence of a β-lactam antibiotic, prevents the transcription of the blaP gene, which encodes the BlaP β-lactamase. In the presence of an antibiotic, BlaI is inactivated and the BlaP β-lactamase is induced [1,2]. The model which has been proposed to explain the induction of BlaP β-lactamase in B. licheniformis 749/I is summarized in Figure 1 [3]. BlaI is a cytoplasmic 128-amino-acid protein composed of two distinct domains: an N-terminal domain (BlaI-NTD: residues 1–82), which contains the DNA-binding motif, and a C-terminal domain (BlaI-CTD: residues 83–128), which is involved in BlaI homodimerization [8]. The dissociation constant (Kd) of the purified BlaI dimer was estimated to be 25 μM by AU (analytical ultracentrifugation) [9]. DNase footprinting experiments have revealed the presence of three regulatory regions that are specifically recognized by BlaI [2]. Two adjacent operators are present in the blaP promoter (OP1 and OP2), and one in the blaI blaR1 promoter (OP3). These operators possess a 23-bp-long dyad symmetry, and exhibit the consensus sequence 5′-aaAgTaTTACAtaTGTAagNtTt-3′ [3] (where the upper- and lower-case letters show conserved and non-conserved bases respectively). By using fluorescent band-shift assays, we suggested that the BlaI dimer recognizes its operators with a distinct global dissociation constant of 10−15–10−14 M2 [9].
Figure 1. β-Lactamase induction mechanism in B. licheniformis 749/I.
The B. licheniformis BlaP β-lactamase is inducible by a β-lactam antibiotic (the inducer) [2]. The regulation of β-lactamase production involves three regulatory genes, blaI, blaR1 and blaR2. The first two genes encode a repressor and a penicillin-sensory transducer respectively ([4], but see [4a]), whereas the third gene is not identified yet. (A) In the absence of a β-lactam antibiotic, the BlaI repressor is bound as a dimer to three operator sequences (OP1, OP2 and OP3) located in the intergenic region between blaP and blaI-blaR operon. Bound to DNA, BlaI prevents the transcription of the blaP, blaI and blaR1 genes ([4], but see [4a]), [5]. (B) When a β-lactam antibiotic is added in the medium, the extracellular penicillin-binding domain of BlaR (BlaR-CTD) is acylated by the antibiotic [6], and a signal is transduced through the transmembrane segment resulting in the activation of the intracellular metalloprotease domain by proteolysis [7]. In B. licheniformis, it has been postulated that the activated metalloprotease converts a pro-coactivator into a co-activator, whose final target is BlaI itself. The BlaI dimer complexed with the co-activator is then released from its operator and β-lactamase is produced at high level ([4], but see [4a]). The product of blaR2 is necessary for the induction process; it is postulated that it can be involved in the activation of the intracellular domain of the BlaR receptor or in the production of the pro-coactivator [3].
BlaI can be hydrolysed by papain, yielding the NTD and CTD domains. BlaI-NTD has been purified by ion-exchange chromatography [10,11]. During this purification step, BlaI-CTD is lost. It is supposed that, during the separation process, BlaI-CTD multimerizes and becomes insoluble (C. Vreuls, unpublished work).
The full-length BlaI, as well as BlaI-NTD, has been studied by NMR spectroscopy [11]. The similarities between BlaI and BlaI-NTD 1H/15N-HSQC (heteronuclear single-quantum coherence spectra) show that the N-terminal part constitutes an independent structural domain. The BlaI-NTD structure was solved by NMR, which revealed that BlaI-NTD belongs to the winged-helix proteins subfamily, which are member of the DNA-recognition helix–turn–helix superfamily [12]. On the other hand, all resonances corresponding to the CTD (amino acids 82–128) were broad, poorly resolved and located in the region of the spectra corresponding to an unfolded polypeptide chain. These results suggest that BlaI-CTD is largely unfolded or highly flexible in comparison with the NTD. Recent X-ray crystallographic data obtained with the Staphylococcus aureus MecI repressor, a protein homologous with BlaI, show that MecI forms a dimer composed of two independent winged-helix domains and two intertwining dimerization domains. Each NTD binds a palindromic DNA operator half-site, and each CTD is composed of three α-helices held together by hydrophobic forces (Figure 2) [13,14].
Figure 2. Views of B. licheniformis BlaI-NTD and S. aureus MecI.
(A) B. licheniformis BlaI-NTD (residues 1–82) belongs to the winged-helix subfamily and consists of a three-stranded β-sheet (S1, Ser23–Asn25; S2, Leu57–Glu62; S3, Val65–Pro70) packed against three α-helices (H1, Asp9–Lys20; H2, Thr26–Thr36; H3, Pro41–Lys53) arranged in the order H1-S1-H2-T1-H3-S2-W1-S3 [11]. S2 and S3 form an antiparallel hairpin (loop called wing W1: Gly63–Arg64) and S1 is connected in parallel with S3. T1 (Ser37–Ser40) is a type 1 turn connecting H2 and H3. The α-helices are in yellow the β-sheet is in red and the tryptophan residues are in black. (B) The S. aureus MecI dimer is composed of two independent winged-helix domains and of two intertwining dimerization domains. The overall fold topology of the NTD is H1-H2-H3-S1-W1-S2, and the CTD is composed of three consecutive α-helices H4-H5-H6 held together by two hydrophobic cores between H4 and H6 [13]. The first monomer is in blue. The NTD of the second monomer (residues 1–73) is in the same colours as BlaI-NTD, and the CTD (residues 74–123) is in green. The S. aureus BlaI and MecI repressors are 60% identical when compared with each other, and 31–41% identical when compared with the B. licheniformis BlaI repressor [14].
In order to obtain more structural information about the BlaI CTD, we have investigated the GdmCl (guanidinium chloride)-dependent unfolding pathway of BlaI, and of its separated domains. GdmCl-induced unfolding was monitored by several techniques: intrinsic and ANS (8-anilinonaphthalene-l-sulphonic acid) fluorescence, far- and near-UV CD spectroscopy, cross-linking, AU, SEC (size-exclusion chromatography) and NMR spectroscopy. These experiments have generated complementary data that lead us to propose a multi-step process for BlaI unfolding. All changes were found to be reversible, and the proposed model contains four different steps in which the two BlaI domains sequentially and independently unfold. The implications of these results on the induction process are also discussed.
MATERIALS AND METHODS
Reagents
GdmCl (>99%) and ANS were purchased from Aldrich. Urea (>99%) was from Merck, and CHAPS was from Boehringer. DSP [dithiobis(succinimidyl propionate)] was from Pierce. Ni-NTA (Ni2+-nitrilotriacetate) agarose was obtained from Affiland. Superdex 75 for gel filtration was purchased from Amersham Biosciences. All solutions were prepared with Milli-Q water and filtered through 0.22 μm pore size membranes (Millipore) before use. All others reagents were of analytical grade.
Expression and purification of BlaI and BlaI(His)6
BlaI and BlaI(His)6, a BlaI recombinant protein with a C-terminal hexahistidine tag, were produced and purified as described previously [3,10]. The uniformly 15N-labelled BlaI sample was prepared as described previously [11]. The protein concentrations were determined using the bicinchoninic acid assay (Pierce), and by measuring the absorbance at 280 nm using a molar absorption coefficient of 16600 M−1·cm−1.
Papain digestion and BlaI-NTD and BlaI-CTD(His)6 purification
Purified BlaI(His)6 in buffer A (50 mM phosphate buffer, pH 8.0, in the presence of 500 mM NaCl) was subjected to papain digestion at 28 °C (1% mol/mol) to generate BlaI-NTD (residues 1–82) and BlaI-CTD(His)6 (residues 83–134) [11]. After an overnight incubation, cleavage was complete, and the two fragments were applied to a Ni-NTA column pre-equilibrated in buffer A. Under these conditions, BlaI-NTD was eluted in the void volume, whereas BlaI-CTD(His)6 was retained on the column. The elution of BlaI-CTD(His)6 was achieved by using buffer B (400 mM imidazole, pH 8.0, supplemented with 13 mM CHAPS). BlaI-NTD was dialysed against buffer C (50 mM phosphate, pH 7.6, supplemented with 200 mM KCl) and concentrated by ultrafiltration using a 5000 Da cut-off membrane (Millipore). BlaI-CTD(His)6 was concentrated, and buffer B was exchanged against buffer C supplemented with 13 mM CHAPS by ultrafiltration using a 1000 Da cut-off membrane. Both fragments were characterized by MALDI–TOF MS (matrix-assisted laser-desorption ionization–time-of-flight MS), as described previously [11]. N-terminal sequencing of BlaI-NTD and BlaI-CTD(His)6 was performed using a Procise 492 pulsed liquid-phase protein sequencer (Applied Biosystems) with 20–30 pmol of protein.
Unfolding and refolding studies
Unfolding experiments were performed at room temperature by incubating BlaI, BlaI-NTD and BlaI-CTD(His)6 (at concentrations ranging from 2 μM to 400 μM) in their respective buffers in the presence of various GdmCl concentrations (for further details, see the Figure legends). The GdmCl concentration was determined from refractive index measurements [15] using an Atago R5000 hand refractometer. Refolding studies were initiated by diluting 10 μl of the BlaI, BlaI-NTD or BlaI-CTD(His)6 GdmCl-denatured samples (prepared as above at a 10-fold-higher protein concentration) with 90 μl of appropriate buffer.
Fluorescent band-shift assay
The fluorescent band-shift assays were performed as described previously [16] with the fluorescent, Cy5-labelled double-stranded oligonucleotide OP1 (5′-Cy5-GCATTTAAATCTTACATATGTAATACTTTC-3′).
Fluorescence measurements
Both intrinsic and ANS fluorescence emission spectra were recorded on a PerkinElmer LS50B spectrofluorimeter. Excitation and emission slit widths were 4 and 5 nm respectively, and the scan speed was 350 nm·min−1. Cuvettes with 1 cm path-length were used.
Intrinsic fluorescence measurements were performed using an excitation wavelength of 280 or 295 nm, and the emission spectra were recorded from 300–420 nm. The protein (2 μM) was conditioned in buffer C supplemented with increasing concentrations of GdmCl. The spectra were measured ten times, averaged and corrected by subtraction of the solvent spectrum obtained under identical conditions. Two fluorescence parameters [17] were considered in the present study: the fluorescence intensity at single excitation and emission wavelengths, and the maximum emission wavelength (λmax). The fraction of unfolded protein (FU) at increasing GdmCl concentrations was calculated using the following equation [15]:
 |
where f is the observed fluorescence intensity at a given GdmCl concentration, fU the fluorescence intensity when the protein is completely unfolded and fN the fluorescence intensity of the native protein.
ANS fluorescence measurements were performed using a protein concentration of 25 μM with an excitation wavelength of 370 nm. Emission spectra for BlaI-CTD(His)6 were recorded from 420–600 nm in buffer C supplemented with 13 mM CHAPS. The fluorescence spectra were corrected for the background fluorescence of ANS. The ANS concentration (determined from the molar absorption coefficient of 5000 M−1·cm−1 at 350 nm) was estimated to be 500 μM [18].
CD measurements
CD measurements were performed using a Jobin-Yvon CD6 spectropolarimeter under constant nitrogen flow. In the far-UV region (195–260 nm), spectra were recorded in a 0.1 cm cell at protein concentrations of 0.5 mg·ml−1, whereas in the near-UV region (250–300 nm), a 1 cm cell was used with a protein concentration of 1 mg·ml−1. For BlaI-CTD(His)6 experiments, Buffer C was used supplemented with 13 mM CHAPS. Spectra were acquired at a scan speed of 20 nm·min−1, with a 2 nm bandwidth and 1 s integration time. The spectra were measured five times, averaged and corrected by subtraction of the solvent spectrum obtained under identical conditions. Results are expressed in terms of molar ellipticity ([θ]). The fraction of unfolded protein FU at various GdmCl concentrations was calculated using the following equation [15]:
 |
where θ is the observed molar ellipticity at a given GdmCl concentration, θU is the molar ellipticity when the protein is completely unfolded and θN is the molar ellipticity of the native protein.
Cross-linking experiments
The stock solution of DSP (2 mM) was prepared in DMSO. Cross-linking experiments were performed in buffer C supplemented with the required GdmCl concentration. Protein (20 μmol) was mixed with 50 μmol of DSP in a total volume of 200 μl. The mixture was incubated for 2 h at 25 °C. The excess of free cross-linking reagent was eliminated by adding 10 μl of 1M Tris/HCl, pH 8.0 and incubating for 15 min on ice. GdmCl-denatured and cross-linked samples were then dialysed for 2 h against buffer D [50 mM Hepes, pH 7.6, supplemented with 200 mM KCl, 1 mM EDTA and 5% (v/v) glycerol] and subjected to SDS/PAGE under non-reducing conditions (i.e. without 2-mercaptoethanol) [19].
AU experiments
Sedimentation velocity experiments were performed with a Beckman Optima XLA instrument at 20 °C using optical absorption detection. BlaI and BlaI-NTD concentrations were 0.9 mg/ml in all experiments. For BlaI-OP1 experiments, one equivalent of DNA (OP1) was added for two equivalents of BlaI. Buffer D was supplemented with the appropriate GdmCl concentration. BlaI and BlaI-NTD samples (500 μl) and the buffer against which the protein had been dialysed were placed in cells fitted with double sector centrepieces and quartz windows. Sedimentation was accomplished at a rotor speed of 40000 rev./min. Cells were scanned at 280 nm (or 292, 295 and 298 nm for the BlaI DNA-binding study); 10–20 scans were collected for each cell. The scans were analysed by the method of Stafford [20]. Apparent sedimentation coefficients S were corrected to S20,w for both viscosity and temperature, as described elsewhere [20]. The densities and relative viscosities of the solutions were determined experimentally, and the partial specific volumes of the proteins were calculated from the amino acid sequences of the proteins.
SEC experiments
GdmCl-induced unfolding of BlaI was analysed by SEC using a Superdex 75 pre-packed FPLC column (10 mm×300 mm, gel volume 24 ml; Amersham Biosciences) calibrated as described by Uversky [21]. The mobile phase was buffer D added with the required GdmCl concentration. Before injection, samples (50 μM) were filtered through a 0.22 μm filter (Millipore). Samples were eluted at a flow rate of 1 ml/min. All measurements were made at room temperature. The protein elution profile was monitored by recording the absorbance at 280 nm. Stokes radii (Rs) were estimated from the elution volumes (Ve) according to the equation: 1000/Ve=11.71+2.97 Rs [21].
NMR studies
Two-dimensional 1H/15N-HSQC spectra were recorded at 25 °C on a Varian INOVA 800 spectrometer equipped with a triple resonance (1H, 15N and 13C) probe and shielded gradients. Samples contained 0.4 mM 98% 15N-enriched BlaI or 0.3 mM 98% 15N-enriched BlaI-NTD in buffer C supplemented with the required GdmCl concentration. The initial sample contained no GdmCl and the 1H2O/2H2O ratio was 9:1. GdmCl salt was added progressively in order to obtain concentrations of 1 M, 1.5 M, 2 M, 3 M, 4 M and 5 M. A 1H/15N-HSQC spectrum was recorded at each GdmCl concentration. To verify the reversibility of BlaI and BlaI-NTD unfolding processes, 1H/15N-HSQC spectra of BlaI and BlaI-NTD were recorded after complete removal of the GdmCl by dialysing against buffer C. 1H and 15N chemical shifts are expressed relative to sodium 2.2-dimethyl-2-silapentane-5-sulphonate (DSS) and liquid ammonia respectively. Maximum t1 and t2 values were 128 ms and 154 ms, and the spectral widths were set to 2000 Hz and 6666 Hz for 15N and 1H dimensions respectively. Data processing was performed using the FELIX program version 2000 (Accelrys).
RESULTS
Purification and characterization of BlaI N- and CTDs
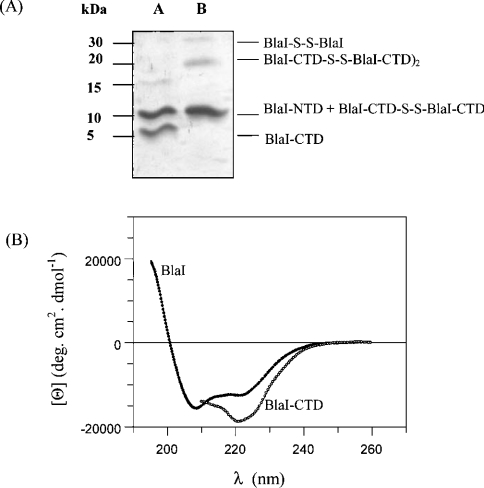
From a papain digest, BlaI-NTD (Met1–Ser82) could easily be purified on an SP Sepharose Fast Flow column [11], whereas BlaI-CTD (His83–Glu128) could not be eluted. This was unexpected on the basis of the pI value of 5.1 computed according to the BlaI-CTD amino acid sequence. A hexahistidine tag was added to the C-terminal end of BlaI in order to purify the CTD [10]. Fragments generated by papain digestion of the purified BlaI(His)6 were applied on to an Ni-NTA affinity column pre-equilibrated in buffer A. As expected, BlaI-NTD was eluted in the void volume, whereas BlaI-CTD(His)6 was adsorbed and subsequently eluted with buffer B, which contained imidazole and 13 mM CHAPS. Addition of a mild zwitterionic detergent (CHAPS) was necessary to prevent BlaI-CTD(His)6 aggregation both upon elution and during all subsequent experiments. The yield of BlaI-CTD(His)6 purification was very poor (5%). N-terminal sequencing and MALDI-TOF experiments revealed that BlaI-NTD extends from amino acids 1–82 (molecular mass 9490 Da) and BlaI-CTD(His)6 from residues 92–134 (molecular mass 4220 Da). MALDI-TOF-MS also highlighted the presence of multimeric forms of the BlaI-CTD(His)6 fragment. These results are in good agreement with those obtained by sedimentation velocity and cross-linking experiments. Indeed, in the stoichiometric mixture of BlaI-NTD and BlaI-CTD present in a papain digest, only BlaI-CTD could be cross-linked by DSP (Figure 3A), and during velocity sedimentation experiments BlaI-NTD behaved as a monomeric protein (results not shown). To verify that BlaI-CTD(His)6 was folded in the presence of CHAPS, far-UV CD spectra were recorded. As shown in Figure 3(B), BlaI-CTD(His)6 exhibits a high percentage of α-helix. The α-helical contents of BlaI, BlaI-NTD and BlaI-CTD(His)6 were calculated as a function of the molar ellipticities at 222 nm (Figure 3B; also see Figures 7A and 7B) [22], and found to be 33%, 25% and 53% respectively. These values are lower than those obtained from the known X-ray structure of the S. aureus MecI repressor (64% for MecI, 53% for MecI-NTD and 81% for MecI-CTD) [13] and from the solution structure of the B. licheniformis BlaI-NTD (43%). MS and CD spectroscopy confirm that BlaI-CTD(His)6 is well folded in the presence of CHAPS, and that it retains its ability to form oligomers. On the other hand, the purified NTD binds its DNA operator with a 500–1000-fold reduced affinity [11], despite the fact that it is no longer able to dimerize, as shown by sedimentation velocity experiments (results not shown).
Figure 3. Characterization of the BlaI CTD.
(A) Study of the oligomerization state of the BlaI CTD by cross-linking experiments. Purified BlaI was subjected to papain digestion. Cross-linking was performed by mixing 20 μmol of the digested protein with 50 μmol of DSP, and the results were analysed by SDS/PAGE (see the Materials and methods section). Lane A corresponds to the digested protein without DSP, and lane B to the digested protein incubated with DSP. Lane B highlights the dimeric (10 kDa) and tetrameric (20 kDa) forms of the BlaI CTD. (B) Far-UV CD spectra of BlaI and BlaI-CTD(His)6. Proteins were incubated in 50 mM phosphate buffer, pH 7.6, supplemented with 200 mM KCl and 13 mM CHAPS for BlaI-CTD(His)6. The protein concentrations used were 0.5 mg·ml−1 in a 0.1 cm cell.
Figure 7. CD studies of BlaI and BlaI-NTD.
Far-UV CD spectra of (A) BlaI and (B) BlaI-NTD under native conditions (50 mM phosphate buffer, pH 7.6, supplemented with 200 mM KCl) and in 5 M GdmCl (‘GdnHCl’) (same buffer). The protein concentration was 0.5 mg·ml−1 in a 0.1 cm cell. The inset shows GdmCl-induced unfolding transition followed at 222 nm. (C) Near-UV CD spectra of BlaI under native conditions and in 5 M GdmCl (same buffers). The protein concentration was 1 mg·ml−1 in a 1 cm cell. The inset shows GdmCl-induced unfolding transition followed at 282 nm.
GdmCl-induced reversible denaturation of BlaI and of its separated fragments
The reversibility of the GdmCl-induced denaturation of purified BlaI, BlaI-NTD and BlaI-CTD(His)6 was studied by several analytical methods and by band-shift assay (for details, see the Materials and methods section). It was shown that, after removal of GdmCl by dilution concentration of the denatured protein: (i) 95% of BlaI and BlaI-NTD intrinsic fluorescence intensity was recovered (results not shown); (ii) BlaI DNA-binding activity was regained, as revealed by band-shift assay experiments (Figure 4); and (iii) the BlaI-CTD(His)6 far-UV CD spectrum (results not shown), as well as the BlaI and BlaI-NTD 1H/15N-HSQC NMR spectra, were fully recovered (see the NMR studies section below).
Figure 4. Study of the reversibility of the BlaI repressor GdmCl-induced denaturation by band-shift assays.
(A) Binding of BlaI to the OP1 target under native conditions (0 M GdmCl). ‘Free’ and ‘Cp’ represent the free DNA target and the DNA–protein complex respectively. The fluorescence is expressed in arbitrary units. DNA probe OP1 (0.5×10−8 M) was incubated with purified BlaI repressor (3.3×10−8 M) and treated as described previously [16]. (B) Binding of BlaI to the OP1 target after denaturation–renaturation of the protein. Protein and DNA concentrations are the same as in (A).
Intrinsic fluorescence of BlaI and BlaI-NTD
The primary structure of BlaI highlights two tryptophan residues at positions 19 and 39, and four tyrosine residues at positions 68, 77, 90 and 116. Only the latter two tyrosine residues are located in BlaI-CTD. Excitation wavelengths of 280 and 295 nm resulted in similar emission spectra, indicating that the tyrosine residues and, hence, the C-terminal part of the protein do not significantly contribute to the fluorescence spectra.
The BlaI unfolding transition was monitored by following the changes of fluorescence emission intensity and of the maximum fluorescence emission wavelength (λmax) in the presence of different GdmCl concentrations. Time-dependent changes in fluorescence emission intensity at increasing GdmCl concentrations showed significant changes within the first 5 min of incubation and no further modification after the next 12 h, suggesting that an incubation time of 2 h was sufficient to achieve equilibrium at any GdmCl concentration (results not shown).
Intrinsic fluorescence emission spectra of native and unfolded proteins are shown in Figure 5(A). The native state is characterized by a λmax at 342 nm. The denaturation process is accompanied by both a decrease in fluorescence intensity and a red-shift of the λmax to 352 nm, which indicates a complete exposure of the tryptophan residues to the aqueous solvent. Figure 5(B) shows the changes in the λmax of BlaI and BlaI-NTD in the presence of increasing GdmCl concentrations, and revealed that BlaI and BlaI-NTD have the same behaviour upon GdmCl unfolding, with their transition midpoints at approx. 4 M GdmCl. Experiments performed in the 1–10 μM protein concentration range gave identical results.
Figure 5. BlaI and BlaI-NTD intrinsic fluorescence emission spectra at various GdmCl concentrations.
Spectra were recorded at 25 °C. Proteins (2 μM) were in 50 mM phosphate, pH 7.6 buffer supplemented with 200 mM KCl. The excitation wavelength was 280 nm. (A) Fluorescence emission spectra of BlaI under native and unfolded conditions (0 M and 5 M GdmCl (‘GdnHCl’) respectively). A.U., arbitrary units. (B) Variation of the λmax of BlaI (○) and BlaI-NTD (•) as a function of GdmCl concentration.
Urea-induced unfolding of BlaI was also monitored by following the changes in intrinsic tryptophan fluorescence intensity at different denaturant concentrations, and showed that urea-induced unfolding took place at approx. 8 M urea (results not shown).
Unfolding of the C-terminal part of the protein, which cannot be monitored by intrinsic fluorescence, was analysed specifically by far-UV CD and ANS fluorescence.
ANS fluorescence
ANS has been widely used as a sensitive reporter of apolar regions in proteins and as a probe for protein non-native, partially unfolded conformations [23–25]. Such intermediates are characterized by the presence of solvent-exposed hydrophobic clusters. The binding of ANS to apolar regions of proteins results in a significant enhancement of ANS fluorescence intensity and in a pronounced blue-shift of the λmax [23].
The existence of partially unfolded intermediates was investigated by monitoring the binding of ANS to BlaI, BlaI-NTD and BlaI-CTD(His)6 in the presence of increasing GdmCl concentrations (Figures 6A, 6B and 6C respectively). As shown in Figure 6(A), a significant binding of ANS to BlaI occurs at 1.5–1.75 M GdmCl, as revealed by a blue-shift of the λmax from 500 to 445 nm, concomitant with a marked increase in ANS fluorescence intensity (results not shown). A further increase in the GdmCl concentration results in the transfer of the ANS molecules from a hydrophobic to a hydrophilic environment. The binding of ANS to BlaI-NTD (Figure 6B) and BlaI-CTD(His)6 (Figure 6C) was also analysed. BlaI-NTD binds ANS at the same GdmCl concentration as BlaI, but the blue-shift of the λmax value is much smaller. On the other hand, BlaI-CTD(His)6 exhibits quite a different ANS-binding curve measured against GdmCl concentration. The binding of ANS to the native protein fragment suggests the presence of solvent-accessible, non-polar clusters in the C-terminal part of the native BlaI protein, as reported for the homologous S. aureus MecI protein [13]. Indeed, the crystallographic 3D (three-dimensional) structure of the dimeric MecI protein reveals the presence of two hydrophobic cavities between the CTDs of each monomer (Figure 2). On the other hand, the variation of ANS binding to BlaI and BlaI-NTD with increasing GdmCl concentrations suggests that BlaI unfolding is accompanied by the accumulation of at least one intermediate with solvent-accessible, non-polar clusters. In order to obtain a further insight into this aspect, the secondary, tertiary and quaternary structures of the protein were studied by CD spectroscopy, gel-filtration chromatography, cross-linking experiments, AU and NMR spectroscopy.
Figure 6. ANS-bound fluorescence emission spectra of BlaI, BlaI-NTD and BlaI-CTD(His)6.
Changes of the emission λmax of ANS bound to (A) BlaI, (B) BlaI-NTD and (C) BlaI-CTD(His)6 as a function of GdmCl (‘GdnHCl’) concentration. Spectra were recorded at 25 °C. Proteins (25 μM) were incubated in 50 mM phosphate buffer, pH 7.6, supplemented with 200 mM KCl and also 13 mM CHAPS for BlaI-CTD(His)6. The ANS concentration was 500 μM. The excitation wavelength was 350 nm.
CD studies
Far-UV CD spectroscopy was used to monitor changes in BlaI and BlaI-NTD secondary structures upon GdmCl-induced unfolding. The far-UV CD spectra of BlaI and BlaI-NTD in their native and unfolded states are shown in Figures 7(A) and 7(B) respectively. Both spectra are very similar, and exhibit strong positive maxima at approx. 195 nm and two negative minima at 208 and 222 nm, typical of proteins containing α-helical secondary structures. Incubation of BlaI in GdmCl solutions results both in changes in the shape of the CD spectra and in a GdmCl-concentration-dependent loss in ellipticity. The changes in BlaI ellipticity values at 222 nm with GdmCl concentration highlight two sharp and well-separated transitions (Figure 7A, inset). The first transition occurs at approx. 1 M GdmCl, and is followed by a plateau between 1 and 3.5 M, whereas the second transition occurs at approx. 4 M GdmCl. Only approx. 15% loss of secondary structure is associated with the first transition, indicating only partial unfolding of BlaI molecules under these conditions. The unfolding transition of the isolated BlaI-NTD fragment monitored by CD at 222 nm is monophasic (Figure 7B, inset) with a transition midpoint identical with that of the second transition of the full-length BlaI.
Intrinsic fluorescence data (Figure 5B) suggest that the BlaI-NTD 3D structure is not modified at GdmCl concentrations below 4 M. Thus the transition monitored by far-UV CD at approx. 1 M GdmCl (Figure 7A) is likely to be due to the unfolding of the C-terminal part of the protein. This hypothesis is in agreement with the ANS fluorescence experiments performed with BlaI-CTD(His)6 (Figure 6C), which show that ANS binds to the native CTD and is completely released after the addition of 1 M GdmCl.
As an alternative probe for tertiary structure changes, the BlaI CD spectrum was also measured in the 250–300 nm range. In the absence of denaturant, the spectrum shows a broad negative band with a minimum at 282 nm (Figure 7C). In the presence of increasing GdmCl concentrations, a progressive decrease in the CD signal is observed between 1–5 M GdmCl (Figure 7C, inset). A possible explanation for this apparent discrepancy between the two optical techniques is that the near-UV CD signal arises from aromatic residues other than those responsible for the intrinsic fluorescence of the protein. Thus the two tyrosine residues (Tyr90 and Tyr116) belonging to the CTD might well contribute significantly to the near-UV CD spectrum of the native BlaI protein, and hence explain how the unfolding transitions observed by fluorescence (Figure 5B) and near-UV CD (Figure 7C, inset) measurements are not superimposable. Note that with both techniques, complete unfolding of the protein is achieved in the presence of 5 M GdmCl.
NMR studies
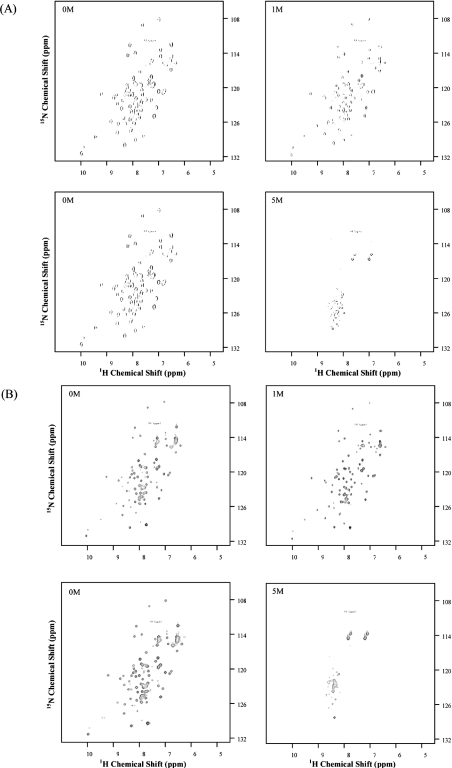
The influence of increasing the GdmCl concentration on the structural changes of BlaI and BlaI-NTD has been monitored by recording 1H/15N-HSQC NMR spectra (Figures 8A and 8B) [26–28]. These NMR spectra highlighted the fact that the BlaI NTD behaves exactly the same upon GdmCl denaturation in the full-length protein or in BlaI-NTD.
Figure 8. Two-dimensional 1H/15N-HSQC NMR spectra of BlaI and BlaI-NTD.
(A) BlaI (0.4 mM) and (B) BlaI-NTD (0.3 mM) were in 50 mM phosphate buffer, pH 7.6, supplemented with 200 mM KCl and the appropriate GdmCl concentration: 0 M GdmCl (upper left panels), 1 M GdmCl (upper right panels) and 5 M GdmCl (lower right panels). The latter spectra (lower left panels) demonstrate the full renaturation of the unfolded proteins after removal of the denaturant (for further details, see the text and the Materials and methods section). A decrease of peak intensity was observed for all 1H/15N cross-peaks upon increasing the GdmCl concentration. This was due to the change in the quality factor of the probe (Q factor) linked to the presence of high amounts of salt [29].
In the BlaI native HSQC spectrum (Figure 8A, top left panel), only the cross-peaks corresponding to the 82 N-terminal amino acids can be assigned [11]. CD studies showed that the CTD contains elements of secondary structure, but the dynamic features are such that it cannot be resolved by NMR spectroscopy [11]. Upon increasing the GdmCl concentration (Figures 8A and 8B, top right panels), chemical shift variations of 0.08 p.p.m. (δH) maximum are observed in solvent-exposed parts of the protein, i.e. residues 6–9, 73–80 (NTD N- and C-terminal ends respectively), 61–63 (Wing 1) and 68 and 69 (β-strand 3). These shifts are due to the modification of the electrostatic environments of these residues caused by an increasing salt concentration. Chemical shifts of residues belonging to the core of the protein remained unchanged up to 4 M GdmCl (Figures 8A and 8B, lower right panels). At 5 M GdmCl, all signals have been replaced by signals characteristic of the denatured state (Figures 8A and 8B, lower right panels). These data show that a total and abrupt denaturation of the N-terminal part occurs for both BlaI and BlaI-NTD between 4 and 5 M GdmCl.
After removal of GdmCl by extensive dialysis against buffer C, the BlaI and BlaI-NTD native spectra are fully recovered (Figures 8A and 8B, lower left panels). These data, combined with the full recovery of the biological activity after an unfolding/refolding cycle, unambiguously demonstrate that the BlaI and BlaI-NTD unfolding processes are fully reversible.
The NMR results confirm that the NTD of the entire BlaI protein is natively folded at 1 M GdmCl, and hence that the transition monitored by far-UV CD spectroscopy at approx. 1 M GdmCl is most likely to be due to the unfolding of the CTD. Conversely, the transition monitored by far-UV CD, intrinsic fluorescence and NMR at approx. 4 M GdmCl is associated with the unfolding of the NTD. However, these results do not supply any information about the oligomerization state of BlaI. To study this phenomenon, three other experiments were performed: cross-linking experiments with DSP [30,31], sedimentation velocity experiments [32] and SEC [33,34].
Cross-linking with DSP and SDS/PAGE analysis
Figure 9(A) shows the electrophoretic behaviour of the BlaI protein after covalent cross-linking with DSP in the presence of various GdmCl concentrations. In the 0–1 M GdmCl concentration range, the tetrameric, dimeric and monomeric forms co-exist. At 2 M GdmCl, both dimeric and monomeric species are present, but the proportion of dimer decreases. At 3 M GdmCl and above, only the monomeric species can be detected.
Figure 9. Effect of GdmCl (‘GdnHCl’) on the oligomerization state of BlaI.
(A) SDS/PAGE after covalent cross-linking of the BlaI protein at increasing GdmCl concentrations. Protein (20 μM) was incubated for 2 h with DSP and subsequently the reaction was quenched 15 min on ice with 10 μl of 1 M Tris, as described in the Materials and methods section. Samples were electrophoresed on an SDS/4–15% acrylamide gel. Shown are standard markers (lane 1), BlaI after cross-linking in the absence of GdmCl (lane 2) and protein pre-incubated in a GdmCl concentration of 1, 2, 3 or 4 M before cross-linking (lanes 3–6 respectively). (B) Sedimentation velocity experiments: GdmCl concentration-dependence of the sedimentation coefficients s20,w (S). BlaI (60 μM) was incubated in 50 mM Hepes buffer, pH 7.6 containing 200 mM KCl, 1 mM EDTA, 5% glycerol and supplemented with the required GdmCl concentration. The samples were centrifuged at 40000 rev./min and scans were collected every 15 min at a wavelength of 280 nm. Sedimentation coefficients were calculated according to the method of Stafford (see [20]).
AU studies
Equilibrium AU experiments were performed in a BlaI concentration range of 10–30 μM, and revealed that monomer, dimer and tetramer co-exist in solution with a [tetramer]/[dimer] ratio equal to 10−5:1 [4]. Sedimentation coefficients (s20,w) determined in the 0–5 M GdmCl concentration range are shown in Figure 9(B). Between 0 and 1.5 M GdmCl, a decrease in the s20,w values was observed, probably due to the release of water bound to the protein. Between 1.5 and 2.5 M, a further decrease in s20,w values occurred, probably corresponding to the dissociation of the dimers into monomers. Above 2.5 M GdmCl, only BlaI monomers were observed. In conclusion, cross-linking and sedimentation velocity experiments reveal that the dissociation of the dimeric protein into monomers takes place at approx. 2.5 M GdmCl.
SEC experiments
The behaviour of BlaI upon gel filtration was analysed under both native and denatured conditions [21,33,34]. Figure 10 shows BlaI elution profiles at different denaturant concentrations. Native BlaI elutes as a single peak with a Ve of 11.2 ml, corresponding to an Rs of 26.1 Å (1 Å=0.1 nm). When the GdmCl concentration is increased to 1.5 M, the peak is shifted to a smaller Ve (10.5 ml). This 10% increase in the Rs value to 28.1 Å suggests a transition to a slightly more expanded compact intermediate state [21]. At approx. 2.5 M GdmCl, the peak is shifted to a higher Ve of 11.9 ml, corresponding to an Rs value of 24.4 Å. This peak should correspond to the monomeric protein, as revealed by cross-linking and sedimentation velocity experiments. The CTD of this monomeric protein should be unfolded, and the N-terminal part ‘native-like’ (Figures 7A and 7B). At 4 M GdmCl and above, the peak is shifted to a smaller Ve, and at 6 M GdmCl, the Ve corresponds to an Rs value of 34.3 Å (Ve=8.8 ml).
Figure 10. SEC elution profiles of BlaI.
BlaI (50 μM) was incubated for 2 h in 50 mM Hepes buffer, pH 7.6, containing 200 mM KCl, 1 mM EDTA, 5% glycerol and supplemented with the required GdmCl (‘GdnHCl’) concentration, and then deposited on to a Superdex 75 column pre-equilibrated with the same GdmCl concentration. Other conditions were as described in the Materials and methods section. (A) Elution profiles of BlaI at different GdmCl concentrations. (B) Rs of BlaI as a function of GdmCl concentration. D, I, M and U represent respectively the native dimeric protein, the ‘ANS-bound’ intermediate state, the partially unfolded monomer and the unfolded BlaI protein.
GdmCl-induced loss of BlaI DNA-binding activity
To highlight the loss of the BlaI DNA-binding property upon GdmCl addition, sedimentation velocity experiments were performed by mixing 60 μM BlaI with 30 μM OP1 in the appropriate buffer supplemented with increasing GdmCl concentration. Sedimentation coefficients (s20,w) corrected for both viscosity and temperature were determined at three different wave-lengths (292, 295 and 298 nm), and at 14 different denaturant concentrations. The DNA-binding capacity of BlaI was progressively lost between 0.75 and 1.75 M GdmCl (results not shown).
DISCUSSION
Previous NMR studies suggested that the CTD of the BlaI repressor was not well folded, or that it may exhibit a high degree of flexibility [11]. For better characterizing the BlaI CTD, we purified BlaI-CTD(His)6 in the presence of CHAPS to prevent its aggregation. Far-UV CD studies of BlaI-CTD(His)6 highlighted the presence of α-helices in the purified BlaI-CTD domain (Figure 3). The GdmCl-induced unfolding of BlaI monitored by far-UV CD revealed two transitions at 1 and 4 M GdmCl respectively, whereas the far-UV CD GdmCl-induced unfolding of BlaI-NTD revealed one single transition at 4 M GdmCl. Moreover, ANS binding to BlaI-CTD(His)6 showed that the λmax of emission increased with the GdmCl concentration, and reached a constant maximum value at approx. 1 M GdmCl. At this point, it was assumed that the CTD of BlaI was well folded in the native repressor, and that the unfolding of this domain occurred at 1 M GdmCl. Cross-linking, sedimentation velocity and gel-filtration experiments showed that the dimer–monomer transition occurred at 2.5 M GdmCl. On the basis of all the experiments, the unfolding of BlaI can be described by the multi-step pathway depicted by Scheme 1.
Scheme 1. Representation of the GdmCl-induced unfolding pathway of the BlaI repressor.
BlaI denaturation proceeds through the independent sequential unfolding of the two domains and involves three distinct intermediate states: (i) an altered dimer (D') with its CTD in a random state and its NTD in a native conformation; (ii) a partially unfolded ‘ANS-bound dimeric intermediate state’ with highly exposed hydrophobic surfaces; and (iii) a partially unfolded monomer (M) with an intact NTD. It should be noted that the NTD remains folded in all the intermediate states, showing the high stability of this independent domain.
In absence of GdmCl, BlaI is mainly present as a dimer (D state) [9]. After addition of 1 M GdmCl, unfolding of the CTD takes place as shown by comparing the far-UV CD spectra of BlaI and BlaI-NTD (Figures 7A and 7B) and by ANS fluorescence experiments (Figure 6C). The NTD retains its native structure, as shown by NMR experiments (Figure 8B). Cross-linking, sedimentation velocity and gel-filtration experiments show that this first intermediate state (D' state) remains dimeric.
At approx. 1.5–1.75 M GdmCl, the formation of an ‘ANS-bound’ intermediate state occurs. This intermediate is characterized by the exposure of hydrophobic surfaces as indicated by ANS binding (Figure 6A), an increase in the hydrodynamic volume compared with the folded state (Figure 10) and a high content of native-like secondary structure (Figure 7A). On the basis of these experimental results, one cannot be sure that the first and second steps occur successively and not simultaneously. In fact, the small increase in size of the ‘ANS-bound’ intermediate state should be concomitant with the unfolding of the CTD. In this case, the first two intermediate states should be replaced by one single intermediate state.
Further increase in the GdmCl concentration (at approx. 2.5 M) results in the dissociation of the dimeric ‘ANS-bound’ intermediate state into partially unfolded monomers (M state), as shown by cross-linking, sedimentation velocity and gel-filtration experiments. The M state is characterized by a native NTD (see the results for the intrinsic fluorescence and NMR experiments described above) and an unfolded CTD.
At denaturant concentrations above 4 M, intrinsic fluorescence, near- and far-UV CD and NMR spectroscopies showed that BlaI completely loses its secondary and tertiary structures, and exists as an expanded monomer (U state). Moreover, 1H/15N-HSQC NMR spectra and intrinsic fluorescence experiments highlighted the fact that the N-terminal part of BlaI and the isolated BlaI-NTD have exactly the same behaviours upon GdmCl unfolding, i.e. a co-operative and total denaturation of the whole domain. In conclusion, the GdmCl-induced unfolding pathway of the dimeric BlaI repressor occurs in a four-step process. During the first step, the unfolding of the BlaI CTD occurs at approx. 1 M GdmCl, followed in the second step, by the formation of an ‘ANS-bound’ intermediate state at approx. 1.5–1.75 M GdmCl. The third step at approx. 2.5 M GdmCl involves the dissociation of the dimeric intermediate, and generates two partially unfolded monomers. Finally, the unfolding of the BlaI NTD leads to the complete denaturation of the whole protein at approx. 4 M GdmCl.
These results confirm that the N- and C-terminal BlaI domains are totally independent from each other, and show that they are both well-folded domains [11]. Similar sequential, independent unfolding events of distinct domains have been reported for many proteins, such as SpnHL (Streptococcus pneumoniae hyaluronate lyase) [35], papain [36], thermolysin [37], urokinase [38], human interferon-α [39], streptokinase [40], subunit IIABman of the mannose transporter of Escherichia coli [41] and aminoacylase [19].
Whereas the NTD is compact and relatively stable upon denaturation, the CTD is more sensitive to the denaturant conditions and exhibits a larger flexibility. On the basis of the sequence similarities between the CTDs of B. licheniformis BlaI and S. aureus MecI, it can be supposed that BlaI-CTD presents a fold similar to that of MecI-CTD. In particular, most of the hydrophobic residues involved in the C-terminal helix–helix dimeric contacts of the 3D-structure of the MecI repressor are conserved between BlaI and MecI. Garcia-Castellanos et al. [13] reported that two side chains of α-helix 1 (residues Trp13 and Asn17) of each NTD contribute to the dimer stabilization by interacting with helices α4 and α5 of the CTD. These interactions do not exist in BlaI, as shown by the NMR spectra, or by comparing the behaviour of the NTD and CTD fragments to that of the whole protein. Moreover, the residues involved in these interactions are not conserved in BlaI (Trp13 is a leucine residue in BlaI, and Asn17 is a lysine residue). In conclusion, the BlaI CTD must have a fold similar to that of MecI-CTD, but without contacts with the NTD. However, these results do not explain why the (D') intermediate state remains dimeric, although its CTD responsible for dimerization is unfolded. It can be assumed that the dimer is stabilized by hydrophobic interactions between the two unfolded CTDs [42].
As described previously [9], the complex between a BlaI monomer and its DNA target cannot be detected due to the high co-operativity of the binding reaction. By contrast, a BlaI dimer interacts with its DNA target with a high affinity (Kd 4×10−11 M). Moreover, Van Melckebeke et al. [11] have shown that BlaI-NTD, which has lost its ability to dimerize, has a DNA-binding affinity 500–1000 times lower than that of the full-length protein. In the present study, we confirm the importance of the CTD of BlaI for its DNA interaction and for its inactivation during the β-lactamase induction. Indeed, our sedimentation velocity experiments showed that BlaI progressively lost its DNA-binding activity at concentrations of between 0.75 and 1.75 M GdmCl. At 1 M GdmCl, the BlaI CTD is unfolded, and approx. 35% of BlaI DNA-binding activity is lost. At 1.75 M GdmCl, BlaI exists in solution as an ‘ANS-bound’ intermediate state, and has completely lost its high affinity for its DNA target. The two NTDs are probably far from each other, and this prevents the co-operative interactions necessary for the high affinity of BlaI for its DNA target. In conclusion, perturbation of the CTD of BlaI could initiate the disassembly of the BlaI–DNA complex and the loss of the repressor activity. This scenario is in agreement with the hypothesis that a co-activator could destabilize the CTD of the repressor [3]. The high sensitivity to GdmCl and the great flexibility of the CTD reflects thermodynamical and dynamical properties that may be essential for the inactivation of BlaI by the binding of the co-activator during the induction process. In S. aureus, the elimination of the BlaI/MecI dimerization domain by a point proteolytic cleavage results in the inactivation of the S. aureus repressors during the induction process [43]. Examination of the MecI 3D structure shows that the NTD and CTD interact with each other. On the other hand, in B. licheniformis BlaI, NMR experiments highlighted the complete independence of the two domains. Supposing that the contacts between the NTD and CTD of MecI are not crystallographic artefacts, we may surmise that this structural difference between B. licheniformis BlaI and S. aureus MecI could play a role in the different induction processes of these repressors. Identification of the putative co-activator and analysis of its effect on the quaternary structure of BlaI should completely clarify the induction mechanism.
Acknowledgments
This work was supported by the Belgian programme of Interuniversity Poles of Attraction initiated by the Federal Office for Scientific Technical and Cultural Affaires (PAI no. P5/33), the ‘Fonds National de la Recherche Scientifique’ (FNRS, Crédit aux chercheurs no. 1.5202.02 and FRFC no. 2.4530.03 and 2.4545.01) and the ‘Communauté Française de Belgique’ (projet Tournesol 2003, 03/013). We also thank Eric Sauvage for his assistance in drawing Figure 2, Edwin De Pauw for MS analysis. A. M. and B. J. are Research Associates of the National Fond for Scientific Research (FNRS, Belgium).
References
- 1.Charlier P., Coyette J., Dehareng D., Dive G., Duez C., Dusart J., Fonze E., Fraipont C., Frère J. M., Galleni M., et al. Résistance bacterienne aux β-lactamases. Medecine/sciences. 1998;14:544–549. [Google Scholar]
- 2.Joris B., Hardt K., Ghuysen J. M. Induction of β-lactamase and low-affinity penicillin binding protein 2′ synthesis in Gram-positive bacteria. New Compr. Biochem. 1994;27:505–515. [Google Scholar]
- 3.Filée P., Benlafaya K., Delmarcelle M., Moutzourelis G., Frère J. M., Brans A., Joris B. The fate of the BlaI repressor during the induction of the Bacillus licheniformis BlaP β-lactamase. Mol. Microbiol. 2002;44:685–694. doi: 10.1046/j.1365-2958.2002.02888.x. [DOI] [PubMed] [Google Scholar]
- 4.Himeno T., Imanaka T., Aiba S. Nucleotide sequence of the penicillinase repressor gene penI of Bacillus licheniformis and regulation of penP and penI by the repressor. J. Bacteriol. 1986;168:1128–1132. doi: 10.1128/jb.168.3.1128-1132.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Erratum. J. Bacteriol. 1987;169:3392. [Google Scholar]
- 5.Salerno A. J., Lampen J. O. Transcriptional analysis of β-lactamase regulation in Bacillus licheniformis. J. Bacteriol. 1986;166:769–778. doi: 10.1128/jb.166.3.769-778.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duval V., Swinnen M., Lepage S., Brans A., Granier B., Frère J. M., Joris B. The kinetic properties of the carboxy terminal domain of the Bacillus licheniformis 749/I BlaR penicillin-receptor shed a new light on the derepression of β-lactamase synthesis. Mol. Microbiol. 2003;48:1553–1564. doi: 10.1046/j.1365-2958.2003.03520.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H. Z., Hackbarth C. J., Chansky K. M., Chambers H. F. A proteolytic transmembrane signaling pathway and resistance to β-lactams Staphylococcus aureus. Science. 2001;291:1962–1965. doi: 10.1126/science.1055144. [DOI] [PubMed] [Google Scholar]
- 8.Wittman V., Lin H. C., Wong H. C. Functional domains of the penicillinase repressor of Bacillus licheniformis. J. Bacteriol. 1993;175:7383–7390. doi: 10.1128/jb.175.22.7383-7390.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filée P., Vreuls C., Hermann R., Thamm I., Frère J. M., Joris B. Dimerization and DNA binding properties of the Bacillus licheniformis BlaI repressor. J. Biol. Chem. 2003;278:16482–16487. doi: 10.1074/jbc.M210887200. [DOI] [PubMed] [Google Scholar]
- 10.Gabelica V., Vreuls C., Filée P., Duval V., Joris B., De Pauw E. Advantages and drawbacks of nanospray for studying noncovalent protein-DNA complexes by mass spectrometry. Rapid Commun. Mass Spectrom. 2002;16:1723–1728. doi: 10.1002/rcm.776. [DOI] [PubMed] [Google Scholar]
- 11.Van Melckebeke H., Vreuls C., Gans P., Filée P., Llabres G., Joris B., Simorre J. P. Solution structural study of BlaI: implications for the repression of genes involved in β-lactam antibiotic resistance. J. Mol. Biol. 2003;333:711–720. doi: 10.1016/j.jmb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Gajiwala K. S., Burley S. K. Winged helix proteins. Curr. Opin. Struct. Biol. 2000;10:110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Castellanos R., Marrero A., Mallorqui-Fernandez G., Potempa J., Coll M., Gomis-Ruth F. X. Three-dimensional structure of MecI. Molecular basis for transcriptional regulation of staphylococcal methicillin resistance. J. Biol. Chem. 2003;278:39897–39905. doi: 10.1074/jbc.M307199200. [DOI] [PubMed] [Google Scholar]
- 14.Rowland S. J., Dyke K. G. Tn552, a novel transposable element from Staphylococcus aureus. Mol. Microbiol. 1990;4:961–975. doi: 10.1111/j.1365-2958.1990.tb00669.x. [DOI] [PubMed] [Google Scholar]
- 15.Pace C. N. Denaturation and analyses of urea and guanidine hydrochloride curves transition. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 16.Filée P., Delmarcelle M., Thamm I., Joris B. Use of an Alfexpress DNA sequencer to analyze protein–nucleic acid interactions by band shift assay. BioTechniques. 2001;30:1044–1051. doi: 10.2144/01305rr03. [DOI] [PubMed] [Google Scholar]
- 17.Dumoulin M., Conrath K., Van Meirhaeghe A., Meersman F., Heremans K., Frenken G. L., Muyldermans S., Wyns L., Matagne A. Single domain antibody fragments with high conformational stability. Protein Sci. 2002;11:500–515. doi: 10.1110/ps.34602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khurana R., Udgaonkar J. B. Equilibrium unfolding studies of barstar: evidence for an alternative conformation which resembles a molten globule. Biochemistry. 1994;33:106–115. doi: 10.1021/bi00167a014. [DOI] [PubMed] [Google Scholar]
- 19.Bai J. H., Xu D., Wang H. R., Zheng S. Y., Zhou H. M. Evidence for the existence of an unfolding intermediate state for aminoacylase during denaturation in guanidine solutions. Biochim. Biophys. Acta. 1999;1430:39–45. doi: 10.1016/s0167-4838(98)00282-9. [DOI] [PubMed] [Google Scholar]
- 20.Stafford W. F. Methods for obtaining sedimentation coefficient distributions. In: Harding S. E., Rowe A. J., Horton J. C., editors. Analytical Ultracentrifugation in Biochemistry and Polymer Sciences. Cambridge, U.K.: Royal Society of Chemistry; 1992. pp. 359–393. [Google Scholar]
- 21.Uversky V. N. Use of fast protein size exclusion liquid chromatography to study the unfolding of proteins which denature through the molten globule. Biochemistry. 1993;32:13288–13298. doi: 10.1021/bi00211a042. [DOI] [PubMed] [Google Scholar]
- 22.Chang C. T., Wu C. S., Yang J. T. Circular dichroic analysis of protein conformation: inclusion of the beta turns. Anal. Biochem. 1978;91:13–31. doi: 10.1016/0003-2697(78)90812-6. [DOI] [PubMed] [Google Scholar]
- 23.Semisotnov G. V., Rodionova N. A., Razgulyaev O. I., Uversky V. N., Gripas A. F., Gilmanshin R. I. Study of the molten globule intermediate state in protein folding by a hydrophobic fluorescent probe. Biopolymers. 1991;31:119–128. doi: 10.1002/bip.360310111. [DOI] [PubMed] [Google Scholar]
- 24.Stryer L. The interaction of naphthalene dye with apomyoglobin and apohemoglobin. A fluorescent probe of non-polar binding sites. J. Mol. Biol. 1965;13:482–495. doi: 10.1016/s0022-2836(65)80111-5. [DOI] [PubMed] [Google Scholar]
- 25.Ptitsyn O. B. Molten globule and protein folding. Adv. Protein Chem. 1995;47:83–229. doi: 10.1016/s0065-3233(08)60546-x. [DOI] [PubMed] [Google Scholar]
- 26.Assfalg M., Banci L., Bertinin I., Turano P., Vasos P. R. Superoxide dismutase folding/unfolding pathway: role of the metal ions in modulating structural and dynamical features. J. Mol. Biol. 2003;330:145–158. doi: 10.1016/s0022-2836(03)00533-3. [DOI] [PubMed] [Google Scholar]
- 27.Van Nuland N. A., Meijberg W., Warner J., Forge V., Scheek R. M., Robillard G., Dobson C. M. Slow cooperative folding of a small globular protein HPr. Biochemistry. 1998;37:622–637. doi: 10.1021/bi9717946. [DOI] [PubMed] [Google Scholar]
- 28.Desmadril M., Yon J. M. Evidence for intermediates during unfolding and refolding of a two-domain protein, phage T4 lysozyme: equilibrium and kinetic studies. Biochemistry. 1984;23:11–19. doi: 10.1021/bi00296a003. [DOI] [PubMed] [Google Scholar]
- 29.Gadian D. G. Oxford: Clarendon Press; 1984. Nuclear Magnetic Resonance and its Application in Living Systems. [Google Scholar]
- 30.Mendoza-Hernandez G., Minauro F., Rendon J. L. Aggregation, dissociation and unfolding of glucose dehydrogenase during urea unfolding. Biochim. Biophys. Acta. 2000;1478:221–231. doi: 10.1016/s0167-4838(00)00025-x. [DOI] [PubMed] [Google Scholar]
- 31.Hermann R., Jaenicke R., Rudolph R. Analysis of the reconstitution of oligomeric enzymes by cross-linking with glutaraldehyde: kinetics of reassociation of lactic dehydrogenase. Biochemistry. 1981;20:5195–5201. doi: 10.1021/bi00521a015. [DOI] [PubMed] [Google Scholar]
- 32.Behal R. H., Debuysere M. S., Demeler B., Hansen J. C., Olson M. S. Pyruvate dehydrogenase multienzyme complex. Characterization of assembly intermediates by sedimentation velocity analysis. J. Biol. Chem. 1994;269:31372–31377. [PubMed] [Google Scholar]
- 33.Ackers G. K. Analytical gel chromatography of proteins. Adv. Protein Chem. 1970;24:343–446. doi: 10.1016/s0065-3233(08)60245-4. [DOI] [PubMed] [Google Scholar]
- 34.Uversky V. N., Ptitsyn O. B. “Partly folded” state, a new equilibrium state of protein molecules: four-state guanidinium chloride-induced unfolding of β-lactamase at low temperature. Biochemistry. 1994;33:2782–2791. doi: 10.1021/bi00176a006. [DOI] [PubMed] [Google Scholar]
- 35.Akhtar M. S., Bhakuni V. Streptococcus pneumoniae hyaluronate lyase contains two non-cooperative independent folding/unfolding structural domains: characterization of functional domain and inhibitors of enzyme. J. Biol. Chem. 2003;278:25509–25516. doi: 10.1074/jbc.M301894200. [DOI] [PubMed] [Google Scholar]
- 36.Sharma Y. V., Jagannadham M. V. N-terminal domain unfolds first in the sequential unfolding of papain. Protein Pept. Lett. 2003;10:83–90. doi: 10.2174/0929866033408327. [DOI] [PubMed] [Google Scholar]
- 37.Corbett R. J., Ahmad F., Roche R. S. Domain unfolding and the stability of thermolysin in guanidine hydrochloride. Biochem. Cell. Biol. 1986;64:953–961. doi: 10.1139/o86-127. [DOI] [PubMed] [Google Scholar]
- 38.Bogusky M. J., Dobson C. M., Smith R. A. Reversible independent unfolding of the domains of urokinase monitored by 1H NMR. Biochemistry. 1989;28:6728–6735. doi: 10.1021/bi00442a028. [DOI] [PubMed] [Google Scholar]
- 39.Skamlova Z., Kontsekova E., Kontsek P. Different stabilities of the N- and C-terminal domains of human interferon alpha. Immunol. Invest. 1997;26:453–458. doi: 10.3109/08820139709022701. [DOI] [PubMed] [Google Scholar]
- 40.Conejero-Lara F., Parrado J., Azuaga A. I., Smith R. A., Ponting C. P., Dobson C. M. Thermal stability of the three domains of streptokinase studied by circular dichroism and nuclear magnetic resonance. Protein Sci. 1996;5:2583–2591. doi: 10.1002/pro.5560051221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Markovic-Housley Z., Cooper A., Lustig A., Flukiger K., Stolz B., Erni B. Independent folding of the domains in the hydrophilic subunit IIABman of the mannose transporter of Escherichiae coli. Biochemistry. 1994;33:10977–10984. doi: 10.1021/bi00202a017. [DOI] [PubMed] [Google Scholar]
- 42.Kauzmann W. Some factors in the interpretation of protein denaturation. Adv. Protein. Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- 43.Zhang H. Z., Hackbarth C. J., Chansky K. M., Chambers H. F. A proteolytic transmembrane signaling pathway and resistance to β-lactams Staphylococcus aureus. Science. 2001;291:1962–1965. doi: 10.1126/science.1055144. [DOI] [PubMed] [Google Scholar]