Abstract
In the yeast genera Saccharomycopsis and Ascoidea, which comprise the taxonomic order Ascoideales, nuclear genes use a nonstandard genetic code in which CUG codons are translated as serine instead of leucine, due to a tRNA-Ser with the unusual anticodon CAG. However, some species in this clade also retain an ancestral tRNA-Leu gene with the same anticodon. One of these species, Ascoidea asiatica, has been shown to have a stochastic proteome in which proteins contain ∼50% Ser and 50% Leu at CUG codon sites, whereas previously examined Saccharomycopsis species translate CUG only as Ser. Here, we investigated the presence, conservation, and possible functionality of the tRNA-Leu(CAG) gene in the genus Saccharomycopsis. We sequenced the genomes of 23 strains that, together with previously available data, include almost every known species of this genus. We found that most Saccharomycopsis species have genes for both tRNA-Leu(CAG) and tRNA-Ser(CAG). However, tRNA-Leu(CAG) has been lost in Saccharomycopsis synnaedendra and Saccharomycopsis microspora, and its predicted cloverleaf structure is aberrant in all the other Saccharomycopsis species. We deleted the tRNA-Leu(CAG) gene of Saccharomycopsis capsularis and found that it is not essential. Proteomic analyses in vegetative and sporulating cultures of S. capsularis and Saccharomycopsis fermentans showed only translation of CUG as Ser. Despite its unusual structure, the tRNA-Leu(CAG) gene shows evidence of sequence conservation among Saccharomycopsis species, particularly in its acceptor stem and leucine identity elements, which suggests that it may have been retained in order to carry out an unknown nontranslational function.
Keywords: tRNA, genetic code, yeast, proteomics, translation
Significance.
Yeasts in the genus Saccharomycopsis have changed the genetic code they use in their nuclear genes, so that CUG codons are translated as serine instead of leucine. Surprisingly, their genomes contain genes for both a tRNA-Ser and a tRNA-Leu, corresponding to the new and old genetic codes, respectively, both with the same anticodon CAG. All protein translation in Saccharomycopsis uses the tRNA-Ser, so we investigated the role of the enigmatic tRNA-Leu gene. We found that even though it is present in most Saccharomycopsis species, it is nonessential and has structural abnormalities. We suggest that it may have been retained for a function other than translation.
Introduction
The genetic code was initially thought to be an immutable frozen accident (Crick 1968; Osawa 1995), because reassignment of a codon's meaning from one amino acid to another, or to a stop codon, would have a wide-ranging effect similar to mutating every gene in which the codon occurs. Despite this, it is now well established that several codon reassignments have occurred during evolution (Keeling 2016), especially in mitochondrial genomes. Evolutionary reassignments of a codon from one amino acid to another (“sense-to-sense” reassignments) are much rarer than reassignments of stop codons to sense codons. In bacteria, a recent extensive computational screen identified only six probable instances of sense-to-sense reassignment among 250,000 bacterial genomes examined, all of which involved reassignment of arginine codons (CGG, CGA, or AGG) to other amino acids (Met, Gln, or Trp) (Shulgina and Eddy 2021).
Across all eukaryotic nuclear genomes, only three instances of sense-to-sense codon reassignment have been discovered. They all occurred in budding yeasts (subphylum Saccharomycotina), and they all involved reassignment of the codon CUG, which is translated as leucine in the standard genetic code. Each of the three yeast clades that changed its genetic code is now classified as a separate taxonomic order, recognizing that genetic code change is a strong phylogenetic marker because it is a rare evolutionary event (Groenewald et al. 2023). Two yeast clades separately reassigned the codon CUG from Leu to Ser (Krassowski et al. 2018). One clade, originally called the CUG-Ser or CUG-Ser1 clade, is now called the order Serinales (Groenewald et al. 2023). It is a large clade that contains the families Debaryomycetaceae and Metschnikowiaceae, including the pathogen Candida albicans (Kawaguchi et al. 1989; Santos and Tuite 1995; Sugita and Nakase 1999). The second clade, called the CUG-Ser2 clade or order Ascoideales, is much smaller and includes only two genera: Ascoidea and Saccharomycopsis (Krassowski et al. 2018; Mühlhausen et al. 2018; Junker et al. 2019). A third yeast clade, containing Pachysolen and two other genera, reassigned the codon CUG from Leu to Ala and is called the CUG-Ala clade or order Alaninales (Mühlhausen et al. 2016; Riley et al. 2016; Krassowski et al. 2018; Mühlhausen et al. 2018; Groenewald et al. 2023). Each of these three genetic code reassignments was achieved by duplicating an ancestral tRNA-Ser or tRNA-Ala gene (a member of a multigene family), followed by mutation(s) that changed its anticodon to CAG, thereby allowing CUG codons to be translated by a tRNA charged with Ser or Ala. The Serinales and Ascoideales clades changed their genetic codes by duplication and anticodon mutation of two different tRNA-Ser genes, one from the four-codon box and one from the two-codon box of serine codons (UCN and AGY codons, respectively) (Krassowski et al. 2018). Interestingly, serine and alanine aminoacyl synthetases (SerRS and AlaRS) are the only aminoacyl synthetases that do not use nucleotides in the anticodon to recognize a tRNA's identity when charging it (Giegé et al. 1998; Giegé and Eriani 2023), so Ser and Ala tRNAs can function with any anticodon sequence, whereas tRNAs for the other 18 amino acids cannot. For this reason, it is probably easier to reassign sense codons to Ser and Ala than to any other amino acids (Kollmar and Mühlhausen 2017a, 2017b).
Of the three yeast taxonomic orders with genetic code reassignments, the order Ascoideales is the only one in which species appear to contain competing tRNAs corresponding to both the new genetic code (tRNA-Ser(CAG)) and the old genetic code (tRNA-Leu(CAG)) (Krassowski et al. 2018; Mühlhausen et al. 2018; Junker et al. 2019). In contrast, tRNA-Leu(CAG) has been lost in the Serinales and Alaninales. The presence of both types of tRNA gene in the same genome may indicate that the evolutionary process of changing the genetic code is still in progress in the Ascoideales, whereas it has finished in the other two clades. However, the functional role of tRNA-Leu(CAG) in the Ascoideales is not very clear. One species, Ascoidea asiatica, has been discovered to have a stochastic proteome in which CUG codons in mRNAs are translated randomly as either Ser or Leu, so tRNA-Leu(CAG) and tRNA-Ser(CAG) are both functional in this species (Mühlhausen et al. 2018). In contrast, proteomic investigations of four Ascoideales species in the genus Saccharomycopsis (Saccharomycopsis malanga, Saccharomycopsis capsularis, Saccharomycopsis fibuligera, and Saccharomycopsis schoenii) indicated that they translate CUG only as Ser, which suggests that their tRNA-Leu(CAG) gene is nonfunctional or at least not used for translation (Krassowski et al. 2018; Mühlhausen et al. 2018; Junker et al. 2019). The proteomic data for S. malanga contrast with another study that reported that both tRNA-Leu(CAG) and tRNA-Ser(CAG) are transcribed and aminoacylated in this species, although it was not specifically shown that the tRNA-Leu is aminoacylated with leucine (Shulgina and Eddy 2021).
In this study, our goal was to investigate the presence, conservation, and possible function of tRNA-Leu(CAG) genes in Saccharomycopsis species. We sequenced the genomes of strains spanning the whole genus, constructed a phylogenomic tree, and compared their synteny at the tRNA-Leu(CAG) and tRNA-Ser(CAG) gene loci. We deleted the tRNA-Leu(CAG) gene of S. capsularis and found that it is nonessential. We used proteomics to investigate the genetic code used during sporulation in Saccharomycopsis, because we found bioinformatically that sporulation genes are enriched in sites where CUG codons coincide with conserved Leu residues, but we found that CUG is still translated only as Ser during sporulation. Our results show that although the tRNA-Leu(CAG) gene is retained in most Saccharomycopsis species, the tRNA has structural defects, is not used by the ribosome, and is not essential. We suggest that it may have a nontranslational role.
Results
Phylogenomic Tree of the Genus Saccharomycopsis
We analyzed genome sequence data from 20 species (33 strains) in the genera Saccharomycopsis and Ascoidea, comprising 23 strains that were newly sequenced for this study using Illumina short-read sequencing, and ten strains whose genome sequences were downloaded from the National Center for Biotechnology Information (NCBI) (supplementary table S1, Supplementary Material online). The data set includes almost all the known species in both genera. The average genome assembly size is 16.3 Mb, with a range from 12.2 to 22.3 Mb (supplementary table S1, Supplementary Material online). We did not include known hybrid strains with double-size genomes, such as S. fibuligera strain KJJ81 (Choo et al. 2016), in the analysis. To investigate the phylogenetic relationships among the 33 strains, we identified a set of 1,227 single-copy orthologous protein groups by using OrthoFinder (Emms and Kelly 2019) and constructed a tree by maximum likelihood (see Materials and Methods). The tree (Fig. 1a) was rooted by assuming that Saccharomycopsis is monophyletic and Ascoidea is an outgroup to it. The OrthoFinder analysis, based on gene duplication events, also supported this position for the root.
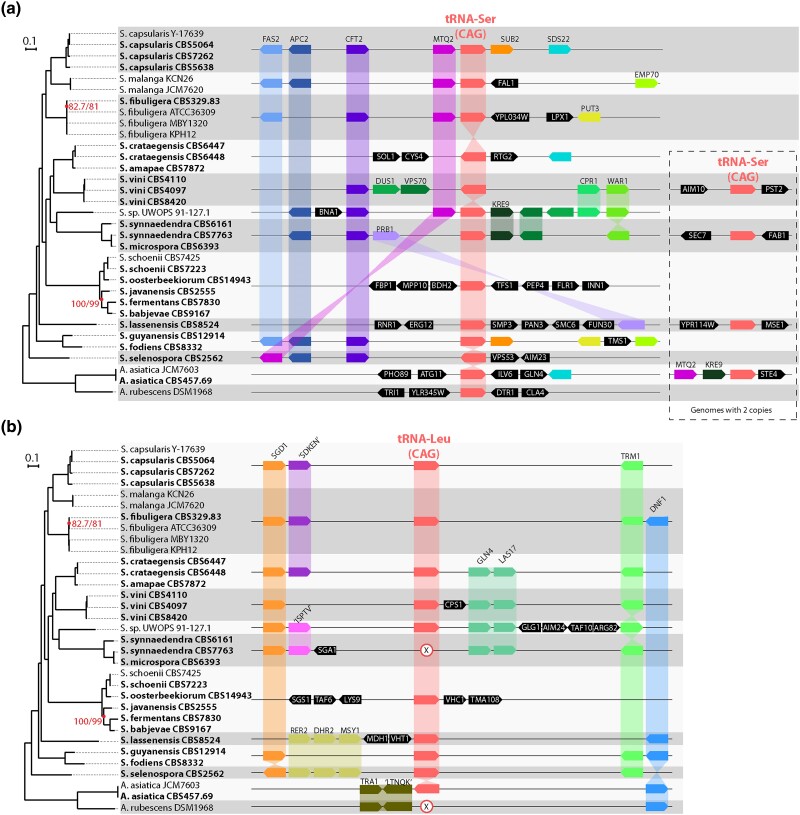
Fig. 1.
Phylogenomic tree of Saccharomycopsis and Ascoidea species, and synteny relationships around a) the tRNA-Ser(CAG) genes and b) the tRNA-Leu(CAG) genes. The same tree is shown in both panels. Strain names are indicated beside species names, and genomes sequenced in this study are indicated in bold. All branches except two were supported by 100% ultrafast bootstrapping and 100% SH-ALRT; the two branches not fully supported are indicated. a) Synteny relationships around tRNA-Ser(CAG) loci. Gray and white horizontal bands indicate groups of strains or species with identical gene arrangements, and only one gene diagram is shown for each group. Some genomes contain a second tRNA-Ser(CAG) gene, shown in the dashed box. In A. asiatica, one sequenced strain (JCM7603) contains two tRNA-Ser(CAG) genes but the other (CBS457.69) contains only one. The copy that is only in JCM7603 shares neighbors (MTQ2 and KRE9) with Saccharomycopsis species. b) Synteny relationships around the tRNA-Leu(CAG) locus. X symbols mark the absence of a tRNA-Leu(CAG) gene in genomes that have lost this gene. Genes are named according to their Saccharomyces cerevisiae orthologs where possible. Other genes are named by conserved sequence motifs they contain.
The tree confirms several clusters of Saccharomycopsis species that were also apparent in a previous tree (Jacques et al. 2014) based on sequencing a small number of loci (SSU rDNA, D1/D2 region of LSU rDNA, and TEF1). Both analyses have a large cluster consisting of S. schoenii, Saccharomycopsis oosterbeekiorum, Saccharomycopsis javanensis, Saccharomycopsis fermentans, and Saccharomycopsis babjevae and the monophyletic pairs Saccharomycopsis crataegensis + Saccharomycopsis amapae, Saccharomycopsis synnaedendra + Saccharomycopsis microspora, and Saccharomycopsis guyanensis + Saccharomycopsis fodiens. However, the positions of these groups relative to each other are substantially different between our tree and that of Jacques et al. (2014). Although many of the internal branches in our phylogenomic tree are short (Fig. 1a), they have strong statistical support whereas many internal branches in the rDNA/TEF1 tree had poor support (Jacques et al. 2014). Our phylogenomic data set is much larger than the rDNA/TEF1 data set and is therefore expected to give a more reliable tree. It indicates that the deepest-branching species in the genus is Saccharomycopsis selenospora. It also places the economically important species S. fibuligera (commonly found in rice wine starter cultures) as a sister clade to S. capsularis and S. malanga. When compared to another recent phylogenomic tree constructed from a smaller number of Saccharomycopsis genome sequences (Yuan et al. 2021), our tree differs in the location of the root and in the position of S. fodiens and the unnamed Saccharomycopsis species UWOPS 91-127.1 relative to the other clades, but again, it has much stronger statistical support.
We detected one strain that appears to have been misidentified. CBS 7763, which was deposited in culture collections as S. synnaedendra, is very closely related to the type strain of S. microspora (CBS 6393) and groups with it rather than with the type strain of S. synnaedendra (CBS 6161) (Fig. 1a), so CBS 7763, therefore, appears to be a strain of S. microspora. This misidentification was also detected independently by Quintilla et al. (2018) using MALDI-TOF mass spectrometry as a tool for species identification.
The tRNA-Leu(CAG) Gene Is Present in Most Ascoideales Species and Is at an Ancestral Location
We used tRNAscan-SE (Lowe and Eddy 1997) to annotate tRNA genes in each genome sequence, supported by BLASTN searches and manual annotation for tRNA genes with CAG anticodons. The total numbers of annotated tRNA genes in the nuclear genome vary extensively, from 108 in Saccharomycopsis species UWOPS 91-127.1 to 760 in Saccharomycopsis vini (supplementary table S2, Supplementary Material online).
We then examined the local gene order around the tRNA-Ser(CAG) and tRNA-Leu(CAG) gene loci (Fig. 1). All the species and strains examined contain at least one gene for tRNA-Ser(CAG), the novel tRNA that allows translation of CUG codons as Ser (Fig. 1a, right panel). Synteny of the nearby protein-coding genes is partially conserved, notably the genes CFT2 and APC2, which are neighbors of tRNA-Ser(CAG) in several Saccharomycopsis clades. There is a second tRNA-Ser(CAG) gene in four clades, but these genes are at different genomic locations that are not conserved so they appear to be the result of independent gene duplications (Fig. 1a, dashed box).
The gene for tRNA-Leu(CAG), the ancestral tRNA that can potentially translate CUG codons as Leu, is present in 17 of the 20 species so it is broadly conserved (Fig. 1b). It is missing in three species: in the species pair S. synnaedendra and S. microspora (Fig. 1b) and in Ascoidea rubescens as previously reported (Krassowski et al. 2018; Mühlhausen et al. 2018). Synteny of other genes around the tRNA-Leu(CAG) locus is reasonably well conserved, which indicates that the tRNA-Leu(CAG) genes are orthologs, except in the clade containing S. fermentans where rearrangements have occurred. Synteny of neighboring protein-coding genes is also conserved in the three species that have lost the tRNA-Leu(CAG) gene, and we did not find any pseudogene or relic of it. The protein-coding gene TRM1, which is beside or close to tRNA-Leu(CAG) in most Saccharomycopsis species (Fig. 1b), is also located beside tRNA-Leu(CAG) in yeasts in the genera Wickerhamomyces and Cyberlindnera (Krassowski et al. 2018). Wickerhamomyces and Cyberlindnera are in the order Phaffomycetales (Groenewald et al. 2023) and translate CUG as Leu, so the shared adjacency of tRNA-Leu(CAG) to TRM1 suggests that this is the ancestral location of the tRNA gene, i.e. that the tRNA-Leu(CAG) gene in most Saccharomycopsis species is a surviving ortholog of the tRNA-Leu(CAG) gene of Wickerhamomyces and Cyberlindnera, which is functional.
The tRNA-Leu(CAG) Gene Is Nonessential in S. capsularis
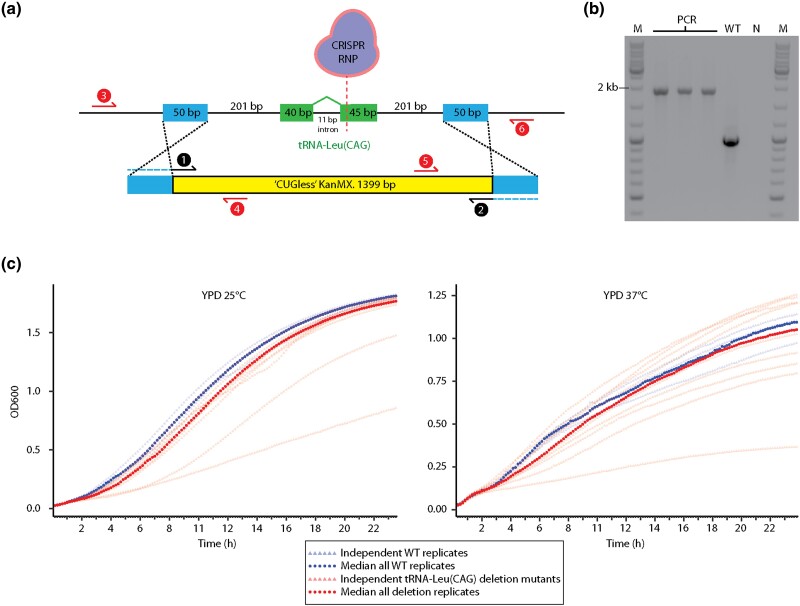
To test whether the tRNA-Leu(CAG) gene is essential in a Saccharomycopsis species, we deleted it by a CRISPR-Cas9 approach in S. capsularis strain NRRL Y-17639. We used an approach similar to Grahl et al. (2017), in which ribonucleoproteins (RNPs) containing Cas9 protein and a tracrRNA:crRNA duplex were cotransformed into competent cells together with a repair template. We first tested the system by disrupting the ADE2 gene of S. capsularis NRRL Y-17639 with kanamycin, nourseothricin, and hygromycin drug resistance cassettes (supplementary fig. S1, Supplementary Material online).
We then used the same approach to delete the tRNA-Leu(CAG) gene, including ∼200-bp upstream and downstream, in the wild-type S. capsularis NRRL Y-17639 genetic background (Fig. 2a). Kanamycin-resistant transformants, from two independent experiments, were verified by PCR (Fig. 2b) and Sanger sequencing of the entire recombinant locus. Nine tRNA-Leu(CAG) deletion mutants were obtained. Transformants were viable and there were no obvious morphological differences (of colonies or of cells examined by light microscopy) between the deletion strains and the wild-type strain, on YPD or synthetic complete media. In liquid growth assays, there was no significant difference between deletion and wild-type strains at either 25 °C or 37 °C, which are respectively the optimal and maximal growth temperature for S. capsularis (Kurtzman et al. 2011) (Fig. 2c). Growth rates of the deletion mutants showed a high variance, which was possibly caused by off-target mutations. However, we cannot rule out the possibility that a phenotype for the deletion strains might be detectable in other growth conditions or by more sensitive assays.
Fig. 2.
Deletion of tRNA-Leu(CAG) in S. capsularis is viable. a) Scheme for deletion of the tRNA-Leu(CAG) gene of S. capsularis NRRL Y-17639 by CRISPR-Cas9 RNPs. The repair template containing a kanamycin resistance cassette (KanMX, modified to lack CUG codons) was amplified from a plasmid using primers with 50-nucleotide tails to generate homology arms matching the regions upstream and downstream of the tRNA-Leu(CAG) gene. CRISPR RNPs containing a guide RNA targeting exon 2 of the tRNA-Leu(CAG) gene were assembled in vitro and electroporated into competent cells. Transformants were screened by junction PCR using primer pairs 3/4 and 5/6 (EOC29/EOC1 and EOC3/EOC30). b) PCR confirmation of three tRNA-Leu(CAG) deletion strains, by amplification of the entire recombinant locus with primer pair 3/6 (EOC29/EOC30). The three lanes marked PCR are transformant colonies with the deletion; WT is a wild-type NRRL Y-17639 control; N is a negative control; and M is a size marker ladder. c) Growth curves comparing the nine independent tRNA-Leu(CAG) deletion strains (red), to three biological replicates of the wild-type strain (blue), at 25 °C and 37 °C. Median values at each time point are shown in the darker shade.
Proteomics of Sporulating Saccharomycopsis Cultures Shows Only CUG-Ser Translation
We hypothesized that Saccharomycopsis cells might change their genetic code during their life cycle or in different growth conditions, with tRNA-Ser(CAG) being active at some stages and tRNA-Leu(CAG) being active at other stages, and all previous proteomic experiments to examine the genetic code in Saccharomycopsis have used only cultures grown in standard laboratory rich media (Krassowski et al. 2018; Mühlhausen et al. 2018; Junker et al. 2019). Our hypothesis was motivated by a bioinformatic analysis that indicated that Saccharomycopsis genes involved in meiosis or sporulation are enriched in CUG codons that align with relatively well-conserved leucine residues in other strains or species (supplementary text S1 and figs. S2 and S3, Supplementary Material online). We therefore conducted proteome analysis by tandem mass spectrometry of cultures of two species, S. capsularis and S. fermentans, grown in sporulation conditions. However, these experiments detected translation of CUG codons only as Ser, in both sporulating and vegetative cultures (supplementary text S1, fig. S4, and table S3, Supplementary Material online).
tRNA-Leu(CAG) Has Structural Irregularities in All Saccharomycopsis Species
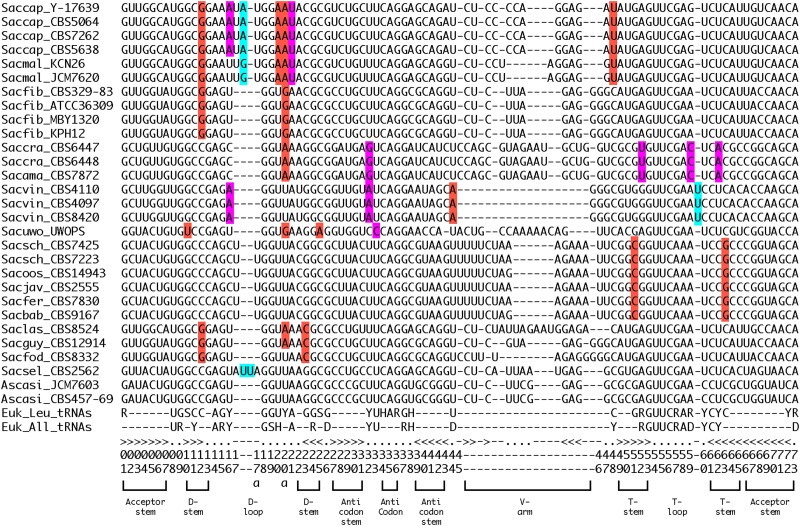
The finding that Saccharomycopsis species appear to translate CUG codons only as Ser, even during sporulation, prompted us to examine the tRNA-Leu(CAG) and tRNA-Ser(CAG) genes and their predicted tRNA cloverleaf structures in more detail. We found that the tRNA-Leu is much more poorly conserved in sequence than the tRNA-Ser, with only 22 completely conserved nucleotide sites as compared to 42 (supplementary fig. S5a, Supplementary Material online). The high diversity of tRNA-Leu(CAG) sequences is also apparent in a phylogenetic tree (supplementary fig. S5b, Supplementary Material online).
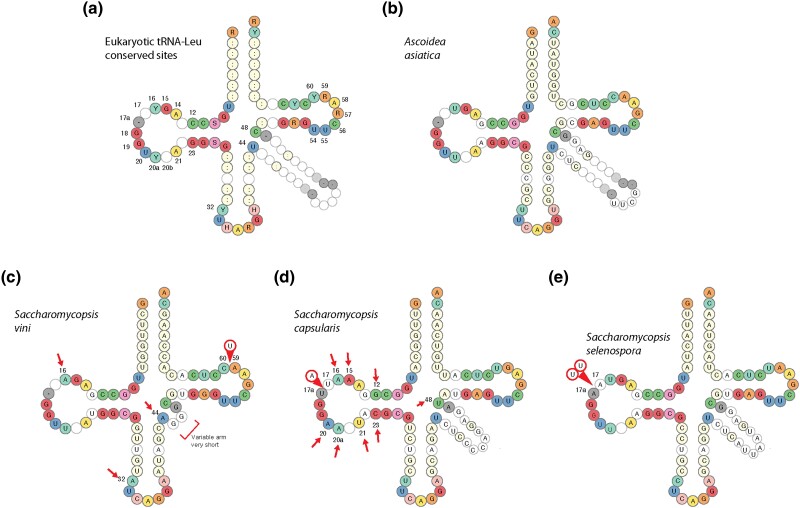
Examination of the predicted cloverleaf structures of tRNA-Leu(CAG) genes revealed unusual features in almost every species (Figs. 3 and 4), when compared to data compiled in the tRNAviz database of tRNA structures from >1,500 species (Lin et al. 2019). Whereas the functional tRNA-Leu(CAG) of A. asiatica has a cloverleaf structure typical of a eukaryotic tRNA-Leu and contains the correct nucleotide at every site that is highly conserved among eukaryotic leucine tRNAs (Fig. 3a and b), the structures predicted in all the Saccharomycopsis species have irregularities that make us doubt that they are functional tRNAs. First, there is no variable arm in tRNA-Leu(CAG) of S. vini (Fig. 3c), whereas an extended variable arm (≥5 bp) is a hallmark of tRNA-Leu, but not of most other tRNA types. Second, some of the predicted Saccharomycopsis tRNA-Leu(CAG) molecules have unusually large D-loops. In A. asiatica, the D-loop is 8 nt long, corresponding to positions 14 to 21 in the standard tRNA numbering scheme (Fig. 3a and b; it also has a mismatch of G13:A22 at the end of the D-stem, as is common in Saccharomycotina tRNA-Leu). The A. asiatica D-loop is consistent in size and sequence with the majority of other eukaryotic tRNA-Leu sequences (Lin et al. 2019). However, the D-loop has expanded to 11 nt in S. capsularis (Fig. 3d) and S. malanga and to 12 nt in S. selenospora (Fig. 3e), making it larger than any conventional eukaryotic tRNA D-loop. The S. capsularis D-loop is also unusual by lacking a uracil residue at position 20; this position normally contains one of the dihydrouracil residues that give the D-loop its name. Third, the predicted Saccharomycopsis tRNA-Leu(CAG) molecules have mutations at positions that are very highly conserved among eukaryotic tRNA-Leu molecules, as shown by the red arrows in Fig. 3. In fact, inspection of the gene sequences in all 16 Saccharomycopsis species that contain tRNA-Leu(CAG) candidates shows that they all contain mutations at positions that are almost universally conserved among eukaryotic tRNA-Leu molecules, and most of them contain multiple mutations of this type (Fig. 4). The A. asiatica tRNA-Leu(CAG) is therefore exceptional in retaining these conserved nucleotides and in retaining translational functionality. However, all the Saccharomycopsis genes still retain the three critical leucylation identity elements A35 (the center of the anticodon), G37 (in the anticodon loop), and A73 (the discriminator position in the acceptor stem) (Soma et al. 1996).
Fig. 3.
Predicted cloverleaf structures of tRNA-Leu molecules. a) Nucleotides that are almost universally conserved in eukaryotic tRNA-Leu molecules, from data compiled in the tRNAviz database (Lin et al. 2019). Numbering is shown for some positions, following the standard tRNA numbering scheme (Sekulovski and Trowitzsch 2022). IUPAC ambiguity codes are used. b to e) Predicted cloverleaf structures of tRNA-Leu(CAG) molecules in A. asiatica and three Saccharomycopsis species. Arrows indicate positions that do not match the highly conserved sites shown in a), and circled bases indicate insertions of bases relative to the standard tRNA structure. Ascoidea asiatica is the only Ascoideales species that retains all the conserved sites and has no insertions.
Fig. 4.
Alignment of tRNA-Leu(CAG) sequences from all sequenced Saccharomycopsis and Ascoidea species, highlighting variants that may make these genes nonfunctional. Standard tRNA position numbering and secondary structure elements are shown at the bottom. Magenta highlighting shows sites that disagree with nucleotides that are strongly conserved in all eukaryotic tRNAs (Euk_All_tRNAs), and red highlighting shows sites that disagree with nucleotides that are strongly conserved in all eukaryotic leucine tRNAs (Euk_Leu_tRNAs), according to data compiled from thousands of genomes in the tRNAviz database (Lin et al. 2019). Blue highlighting shows insertions of bases in the D-loop and T-loop in some species, making these loops longer than can be accommodated by the standard numbering system.
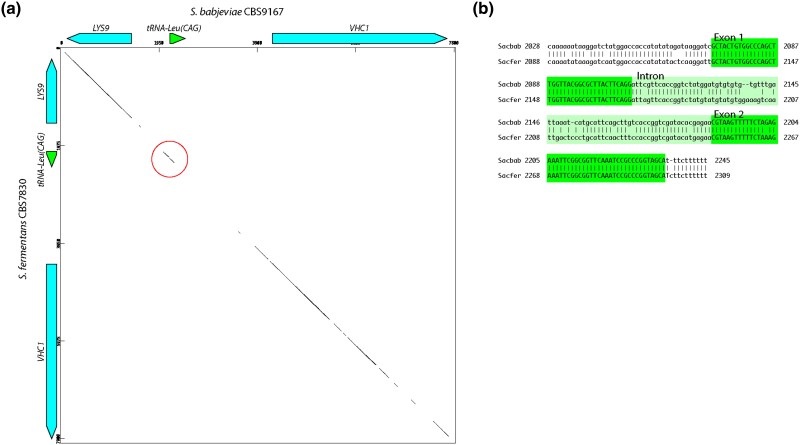
Despite the apparent defects in the cloverleaf structures, comparative analysis indicates that the sequences of the tRNA-Leu(CAG) locus have been preserved during evolution to a greater extent than expected by chance. For example, a dot matrix comparison between S. babjevae and S. fermentans shows that the tRNA locus has been conserved whereas most of the intergenic region that surrounds it (between the neighboring protein-coding genes) has diverged (Fig. 5a). The exons of the tRNA-Leu(CAG) gene have 98.8% sequence identity (only 1 nucleotide different), whereas the intron and the alignable region upstream of exon 1 have diverged (77.8% and 83.7% identity, respectively; Fig. 5b), which is the pattern expected if the gene is being conserved by natural selection. Similarly, the tRNA-Leu(CAG) gene is better conserved than its flanking regions in a comparison among S. capsularis, S. malanga, and S. fibuligera (Krassowski et al. 2018).
Fig. 5.
Conservation of the tRNA-Leu(CAG) locus between S. babjevae and S. fermentans. a) Dot matrix plot of the region containing the tRNA-Leu(CAG) locus and the neighboring protein-coding genes LYS9 and VHC1. Dots indicate regions with ≥25 matches in a 30-bp sliding window. The match at the tRNA locus is circled. b) BLASTN alignment corresponding to the region circled in a). The exons of the tRNA gene are better conserved (only 1 nt different, i.e. 98.8% identity) than the intron (77.8% identity) and the alignable region upstream of the gene (83.7% identity). Regions further upstream and downstream cannot be aligned, as shown by the dot matrix plot.
Discussion
To reconcile the conflicting observations about tRNA-Leu(CAG), we suggest that this gene is losing its role in translation in the Ascoideales. Its role in translation is being replaced by tRNA-Ser(CAG), and this process of replacement has proceeded to different extents in different species; it has proceeded further in Saccharomycopsis and in A. rubescens than in A. asiatica. Further, we suggest that the tRNA-Leu(CAG) gene may have an unknown nontranslational function that has led to the evolutionary retention of the gene in most species while allowing its sequence to diverge to a greater extent than is normal for a tRNA gene. If this nontranslational function requires transcription of the locus, it can explain the transcription and splicing that has been observed (Krassowski et al. 2018; Shulgina and Eddy 2021), and it can explain the greater conservation of the exons than the intron (Fig. 5). If the nontranslational function also requires aminoacylation of the transcript, it can explain the aminoacylation that was detected in S. malanga (Shulgina and Eddy 2021).
Many nontranslational roles have been identified for tRNAs (Katz et al. 2016; Avcilar-Kucukgoze and Kashina 2020). An interesting precedent for what may have happened to the yeast tRNA-Leu(CAG) has been found in a tRNA-Glu gene in plant chloroplast genomes. As well as its role in the translation of GAA codons, chloroplast tRNA-Glu(UUC) is also required in the pathway that plants use for the biosynthesis of tetrapyrroles such as heme and chlorophyll (Barbrook et al. 2006; Agrawal et al. 2020; Kořený et al. 2022). One of the first steps in this pathway takes place in the chloroplast and uses a glutamate residue, which must be attached (charged) to tRNA-Glu(UUC), as a substrate. In photosynthetic plants, the chloroplast genome normally contains 30 different tRNA genes, but in the much-reduced plastid genome of the nonphotosynthetic parasitic plant Balanophora, all the tRNA genes have been lost except for a degenerated tRNA-Glu(UUC) gene (Su et al. 2019). The Balanophora tRNA-Glu(UUC) is predicted to have a badly damaged structure, and it cannot form a normal anticodon loop, but its acceptor stem region and most of the glutamate identity elements are intact. It has been proposed that the RNA produced by this gene is aminoacylated with glutamate, which is then used for tetrapyrrole synthesis, and that it has no translational function (Su et al. 2019).
We suggest that Saccharomycopsis tRNA-Leu(CAG) may have survived because it has an analogous but unknown nontranslational function that requires its transcription, splicing, and aminoacylation. Notably, the acceptor stem of tRNA-Leu(CAG) is well conserved in Saccharomycopsis (Figs. 3 and 4), so it seems possible that the gene's transcript, which retains the three key leucylation identity elements A35, G37, and A73, is recognized and charged by leucyl-tRNA synthetase (LeuRS). We speculate that the gene could produce a charged tRNA-like molecule that has a nontranslational function, for example as a source of amino acids for the posttranslational modification of proteins by enzymes similar to yeast Ate1 and bacterial Leu/Phe transferase (Tobias et al. 1991; Shrader et al. 1993; Abeywansha et al. 2023). However, if a nontranslational function is the explanation for the tRNA-Leu(CAG) gene's survival, it remains unclear why all three bases of its anticodon have remained completely conserved, when only the central base (A35) is a determinant for leucylation (Giegé et al. 1998; Giegé and Eriani 2023).
Materials and Methods
Genome Sequencing
We sequenced 23 strains of Saccharomycopsis by using Illumina technology, including the type strains of most of the known species in the genus (supplementary table S1, Supplementary Material online). Strains were purchased from the Westerdijk Institute for Fungal Diversity (CBS Collection, the Netherlands). Cultures of all strains were grown in YPD at 30 °C overnight. Cells were harvested by centrifugation, and cell pellets were resuspended in 200-µl extraction buffer (2% Triton X-100, 100 mM NaCl, 10 mM Tris pH 7.4, 1 mM EDTA, and 1% SDS) in a 1.5-ml screw-cap tube. Approximately 0.3-g acid-washed glass beads (425 to 600 μm) were added with 200-μl phenol/chloroform/isoamyl alcohol (25:24:1). The mixture was agitated on a 600 MiniG bead beater (Spex SamplePrep) at 1,500 rpm, 4 to 6 times for 30 s each, and centrifuged at 15,000 rcf for 10 min. The top aqueous layer was transferred to a new 1.5-ml screw-cap tube, 200-μl TE buffer was added, and 200 μl of the phenol/chloroform/isoamyl alcohol mixture was added. This was agitated as before on the 600 MiniG bead beater, centrifuged at 14,000 rpm for 10 min. The top aqueous layer was transferred to a new microfuge tube, after which 80-μl 7.5 M ammonium acetate and 1-ml 100% isopropyl alcohol were added to precipitate the DNA. DNA was pelleted by centrifugation at 14,000 rpm, washed using 70% ethanol and dried in a SpeedVac (Eppendorf Concentrator 5301 at 45 °C for 2-min pulses until dry). Pellets were resuspended in 400-µl TE buffer with 1-µl RNase A (10 mg/ml) and incubated overnight at 37 °C. DNA was reprecipitated and washed once more as above and resuspended in 150-µl water. DNA quality and concentration were assessed by gel electrophoresis, NanoDrop, and Qubit measurement. Genomic DNA was sequenced by BGI Tech Solutions (Hong Kong) using an Illumina X Ten instrument (150-bp paired ends, 1.5-Gb raw data per sample).
Genome Assembly, Annotation, and Phylogenomics
Paired-end reads for the 23 strains of this study were quality assessed by FASTQC before and after trimming by Skewer v.0.2.2 (Jiang et al. 2014). De novo genome assemblies were generated by SPAdes v.3.11 (Bankevich et al. 2012) for each set of reads. Genome quality was assessed by QUAST and coverage-versus-length plots (Douglass et al. 2019). Ten other genome sequences were downloaded from NCBI (supplementary table S1, Supplementary Material online). For each of the 33 genome sequences, AUGUSTUS v3.5.0 (Stanke et al. 2008) was used to predict ORFs, and tRNAscan-SE v2.0.5 (Lowe and Eddy 1997) was used to annotate tRNA genes. For some Saccharomycopsis species, the tRNA-Leu(CAG) gene was not identified by tRNAscan-SE and instead was annotated manually based on BLASTN search results. All predicted tRNA genes with CAG as the anticodon were manually inspected and classified as either Ser or Leu tRNAs using identity determinants specific to tRNA-Leu or tRNA-Ser molecules (Giegé et al. 1998; Giegé and Eriani 2023) as well as synteny conservation.
For phylogenomic analysis, the predicted sets of ORFs from Augustus were trimmed to remove ORFs that were incomplete because they reached the end of a contig. This set of ORFs was translated using the standard genetic code, except that CUG codons were translated as “X.” These 33 sets of translated ORFs from each genome were used as input to OrthoFinder v2.5.2 (Emms and Kelly 2019). The resulting set of all single-copy orthologs (1,227) was used for phylogenomic tree construction. These orthogroups were each aligned using MUSCLE v3.8.31 (Edgar 2004), trimmed using trimAl v1.2 (Capella-Gutierrez et al. 2009) with the -automatic heuristic setting, and concatenated together into a supermatrix containing 567,098 sites. IQ-tree v1.6.12 (Nguyen et al. 2015) was used to first find an appropriate maximum likelihood model for this supermatrix and then to construct a tree using that model (LG + F + R5), performing 1,000 ultrafast bootstraps and 1,000 Shimodaira-Hasegawa Approximate Likelihood Ratio Tests (SH-ALRT) (Shimodaira 2002). The branching order of the tree was inspected manually and compared to the OrthoFinder species tree, which is inferred from gene duplication events.
CRISPR RNP Gene Editing in S. capsularis NRRL Y-17639
We adapted a CRISPR RNP protocol from Grahl et al. (2017) and DiCarlo et al. (2013). Saccharomycopsis capsularis NRRL Y-17639 was inoculated in 5-ml YPD, grown at 30 °C overnight, back-diluted to OD600 0.1 in 50-ml YPD, and grown for 14 h at 30 °C. Cells were harvested by centrifugation at 1,780 rcf at 4 °C. The cells were washed in 10-ml ice-cold sterile double-deionized water, centrifuged as before, and washed again in 25-ml ice-cold electroporation buffer (EB; 1 M sorbitol and 1 mM CaCl2). Cells were then centrifuged, resuspended in lithium acetate/DTT (500 mM/10 mM), and incubated for 30 m at 30 °C. They were then centrifuged, washed a final time in 25-ml EB, and resuspended in 200-μl EB. They were kept on ice until needed, and 40 μl of this mixture was used for each transformation reaction. crRNA and tracrRNA ssRNA oligos from the ALT-R system (Integrated DNA Technologies) were annealed as recommended. The annealed guides were incubated with Cas9p for 5 min at room temperature and then kept on ice until ready. For homology directed repair templates, “CUGless” drug marker cassettes with no CUG codons were amplified from plasmid stocks with 70-mer oligonucleotide primers, with 20 bases matching the ends of the marker and 50 bases matching either directly adjacent to the CRISPR cut site (supplementary fig. S1, Supplementary Material online) or several hundred bases upstream/downstream of the cut site (Fig. 2). Typically 4 × 50-μl PCR reactions were pooled together, purified by spin columns, and eluted in 30-μl water to generate each repair template. The CRISPR RNPs (6.6 μl), 2.5 μg of repair template DNA, and 40 μl of the prepared competent cells were mixed together in a microfuge tube on ice. This mixture was transferred into a 2-mm sterile electroporation cuvette (VWR) chilled on ice and electroporated (Bio-Rad GenePulser Xcell; 2,500 V, 25µF, 200Ω). The electroporated PCR mixture was transferred to 7-ml recovery medium (1:1 YPD:1 M sorbitol), and incubated at 30 °C for 2.5 h. Cells were then spun down, resuspended in 200 μl YPD, and plated to the relevant drugs. For selection, 50-ng/μl G-418 or 10-ng/μl nourseothricin was used.
Correct integration of drug cassettes at the target locus was assessed first by colony PCR of the integration junctions. For S. capsularis colony PCR, a colony was resuspended in 200-μl TE, 200-μl phenol/chloroform/isoamyl alcohol was added with 0.3 g acid-washed glass beads (425 to 600 μm), all in a 1.5-ml screw-cap tube. The mixture was treated in a BeadBeater 4 to 6 times for 30 s each and then centrifuged at 15,000 rcf, after which the top aqueous layer was transferred to a new tube and frozen for future use in PCR reactions (0.5 to 1 μl per 20 to 50 μl PCR reaction). Once each integration junction was detected by PCR, the entire locus was amplified by PCR using external primers and Sanger sequenced to confirm its structure. Sequences of oligonucleotides used as PCR primers are given in supplementary table S4, Supplementary Material online.
Although we successfully used CRISPR RNP gene editing to delete tRNA-Leu(CAG) and disrupt ADE2 in S. capsularis strain NRRL Y-17639, which does not sporulate, we were unsuccessful in attempts to make similar deletions of tRNA-Leu(CAG) in three other strains of S. capsularis that sporulate well (CBS5064, CBS5638, and CBS7262).
Supplementary Material
Acknowledgments
We thank Padraic Heneghan for discussion.
Contributor Information
Eoin Ó Cinnéide, UCD Conway Institute and School of Medicine, University College Dublin, Dublin, Ireland.
Caitriona Scaife, Mass Spectrometry Core Facility, UCD Conway Institute, University College Dublin, Dublin, Ireland.
Eugène T Dillon, Mass Spectrometry Core Facility, UCD Conway Institute, University College Dublin, Dublin, Ireland.
Kenneth H Wolfe, UCD Conway Institute and School of Medicine, University College Dublin, Dublin, Ireland.
Supplementary Material
Supplementary material is available at Genome Biology and Evolution online.
Funding
This work was supported by the European Research Council (ERC) under the European Union's Horizon 2020 Research and Innovation Program [grant agreement no. 789341 (Codekiller) to K.H.W.].
Data Availability
New genome sequences reported in this study are listed in supplementary table S1, Supplementary Material online and have been deposited in the ENA/NCBI/DDBJ databases under BioProject PRJNA977123. Mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the following data set identifiers: PXD044638, PXD044720, PXD044729, PXD044731, PXD044788, PXD044742, PXD044763, PXD044772, PXD044785, PXD044786, PXD044693, and PXD044719.
Literature Cited
- Abeywansha T, Huang W, Ye X, Nawrocki A, Lan X, Jankowsky E, Taylor DJ, Zhang Y. The structural basis of tRNA recognition by arginyl-tRNA-protein transferase. Nat Commun. 2023:14(1):2232. 10.1038/s41467-023-38004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal S, Karcher D, Ruf S, Bock R. The functions of chloroplast glutamyl-tRNA in translation and tetrapyrrole biosynthesis. Plant Physiol. 2020:183(1):263–276. 10.1104/pp.20.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avcilar-Kucukgoze I, Kashina A. Hijacking tRNAs from translation: regulatory functions of tRNAs in mammalian cell physiology. Front Mol Biosci. 2020:7:610617. 10.3389/fmolb.2020.610617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012:19(5):455–477. 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbrook AC, Howe CJ, Purton S. Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci. 2006:11(2):101–108. 10.1016/j.tplants.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. Trimal: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009:25(15):1972–1973. 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo JH, Hong CP, Lim JY, Seo JA, Kim YS, Lee DW, Park SG, Lee GW, Carroll E, Lee YW, et al. Whole-genome de novo sequencing, combined with RNA-seq analysis, reveals unique genome and physiological features of the amylolytic yeast Saccharomycopsis fibuligera and its interspecies hybrid. Biotechnol Biofuels. 2016:9(1):246. 10.1186/s13068-016-0653-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick FH. The origin of the genetic code. J Mol Biol. 1968:38(3):367–379. 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- Dicarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013:41(7):4336–4343. 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass AP, O'Brien CE, Offei B, Coughlan AY, Ortiz-Merino RA, Butler G, Byrne KP, Wolfe KH. Coverage-versus-length plots, a simple quality control step for de novo yeast genome sequence assemblies. G3 (Bethesda). 2019:9(3):879–887. 10.1534/g3.118.200745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004:5(1):113. 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019:20(1):238. 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé R, Eriani G. The tRNA identity landscape for aminoacylation and beyond. Nucleic Acids Res. 2023:51(4):1528–1570. 10.1093/nar/gkad007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998:26(22):5017–5035. 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahl N, Demers EG, Crocker AW, Hogan DA. Use of RNA-protein complexes for genome editing in non-albicans Candida species. mSphere. 2017:2(3):e00218-00217. 10.1128/mSphere.00218-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenewald M, Hittinger CT, Bensch K, Opulente DA, Shen X-X, Li Y, Liu C, LaBella AL, Zhou X, Limtong S, et al. A genome-informed higher rank classification of the biotechnologically important fungal subphylum Saccharomycotina. Stud Mycol. 2023:105(1):1–22. 10.3114/sim.2023.105.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques N, Louis-Mondesir C, Coton M, Coton E, Casaregola S. Two novel Saccharomycopsis species isolated from black olive brines and a tropical plant. Description of Saccharomycopsis olivae f. a., sp. nov. and Saccharomycopsis guyanensis f. a., sp. nov. Reassignment of Candida amapae to Saccharomycopsis amapae f. a., comb. nov., Candida lassenensis to Saccharomycopsis lassenensis f. a., comb. nov. and Arthroascus babjevae to Saccharomycopsis babjevae f. a., comb. nov. Int J Syst Evol Microbiol. 2014:64:2169–2175. 10.1099/ijs.0.060418-0. [DOI] [PubMed] [Google Scholar]
- Jiang H, Lei R, Ding SW, Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014:15(1):182. 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker K, Chailyan A, Hesselbart A, Forster J, Wendland J. Multi-omics characterization of the necrotrophic mycoparasite Saccharomycopsis schoenii. PLoS Pathog. 2019:15(5):e1007692. 10.1371/journal.ppat.1007692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz A, Elgamal S, Rajkovic A, Ibba M. Non-canonical roles of tRNAs and tRNA mimics in bacterial cell biology. Mol Microbiol. 2016:101(4):545–558. 10.1111/mmi.13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Honda H, Taniguchi-Morimura J, Iwasaki S. The codon CUG is read as serine in an asporogenic yeast Candida cylindracea. Nature. 1989:341(6238):164–166. 10.1038/341164a0. [DOI] [PubMed] [Google Scholar]
- Keeling PJ. Genomics: evolution of the genetic code. Curr Biol. 2016:26(18):R851–R853. 10.1016/j.cub.2016.08.005. [DOI] [PubMed] [Google Scholar]
- Kollmar M, Mühlhausen S. How tRNAs dictate nuclear codon reassignments: only a few can capture non-cognate codons. RNA Biol. 2017a:14(3):293–299. 10.1080/15476286.2017.1279785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmar M, Mühlhausen S. Nuclear codon reassignments in the genomics era and mechanisms behind their evolution. Bioessays. 2017b:39(5):1600221. 10.1002/bies.201600221. [DOI] [PubMed] [Google Scholar]
- Kořený L, Oborník M, Horáková E, Waller RF, Lukeš J. The convoluted history of haem biosynthesis. Biol Rev Camb Philos Soc. 2022:97(1):141–162. 10.1111/brv.12794. [DOI] [PubMed] [Google Scholar]
- Krassowski T, Coughlan AY, Shen XX, Zhou X, Kominek J, Opulente DA, Riley R, Grigoriev IV, Maheshwari N, Shields DC, et al. Evolutionary instability of CUG-leu in the genetic code of budding yeasts. Nat Commun. 2018:9(1):1887. 10.1038/s41467-018-04374-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman CP, Fell JW, Boekhout T. The yeasts, a taxonomic study. Amsterdam: Elsevier; 2011. [Google Scholar]
- Lin BY, Chan PP, Lowe TM. tRNAviz: explore and visualize tRNA sequence features. Nucleic Acids Res. 2019:47(W1):W542–W547. 10.1093/nar/gkz438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997:25(5):955–964. 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlhausen S, Findeisen P, Plessmann U, Urlaub H, Kollmar M. A novel nuclear genetic code alteration in yeasts and the evolution of codon reassignment in eukaryotes. Genome Res. 2016:26(7):945–955. 10.1101/gr.200931.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlhausen S, Schmitt HD, Pan K-T, Plessmann U, Urlaub H, Hurst LD, Kollmar M. Endogenous stochastic decoding of the CUG codon by competing Ser- and Leu-tRNAs in Ascoidea asiatica. Curr Biol. 2018:28(13):2046–2057.e5. 10.1016/j.cub.2018.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015:32(1):268–274. 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa S. Evolution of the genetic code. Oxford: Oxford University Press; 1995. [Google Scholar]
- Quintilla R, Kolecka A, Casaregola S, Daniel HM, Houbraken J, Kostrzewa M, Boekhout T, Groenewald M. MALDI-TOF MS as a tool to identify foodborne yeasts and yeast-like fungi. Int J Food Microbiol. 2018:266:109–118. 10.1016/j.ijfoodmicro.2017.11.016. [DOI] [PubMed] [Google Scholar]
- Riley R, Haridas S, Wolfe KH, Lopes MR, Hittinger CT, Göker M, Salamov AA, Wisecaver JH, Long TM, Calvey CH, et al. Comparative genomics of biotechnologically important yeasts. Proc Natl Acad Sci U S A. 2016:113(35):9882–9887. 10.1073/pnas.1603941113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos MA, Tuite MF. The CUG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids Res. 1995:23(9):1481–1486. 10.1093/nar/23.9.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulovski S, Trowitzsch S. Transfer RNA processing—from a structural and disease perspective. Biol Chem. 2022:403(8-9):749–763. 10.1515/hsz-2021-0406. [DOI] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002:51(3):492–508. 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Shrader TE, Tobias JW, Varshavsky A. The N-end rule in Escherichia coli: cloning and analysis of the leucyl, phenylalanyl-tRNA-protein transferase gene aat. J Bacteriol. 1993:175(14):4364–4374. 10.1128/jb.175.14.4364-4374.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulgina Y, Eddy SR. A computational screen for alternative genetic codes in over 250,000 genomes. eLife. 2021:10:e71402. 10.7554/eLife.71402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma A, Kumagai R, Nishikawa K, Himeno H. The anticodon loop is a major identity determinant of Saccharomyces cerevisiae tRNA(Leu). J Mol Biol. 1996:263(5):707–714. 10.1006/jmbi.1996.0610. [DOI] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008:24(5):637–644. 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- Su HJ, Barkman TJ, Hao W, Jones SS, Naumann J, Skippington E, Wafula EK, Hu J-M, Palmer JD, dePamphilis CW. Novel genetic code and record-setting AT-richness in the highly reduced plastid genome of the holoparasitic plant Balanophora. Proc Natl Acad Sci U S A. 2019:116(3):934–943. 10.1073/pnas.1816822116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita T, Nakase T. Non-universal usage of the leucine CUG codon and the molecular phylogeny of the genus Candida. Syst Appl Microbiol. 1999:22(1):79–86. 10.1016/S0723-2020(99)80030-7. [DOI] [PubMed] [Google Scholar]
- Tobias JW, Shrader TE, Rocap G, Varshavsky A. The N-end rule in bacteria. Science. 1991:254(5036):1374–1377. 10.1126/science.1962196. [DOI] [PubMed] [Google Scholar]
- Yuan X, Peng K, Li C, Zhao Z, Zeng X, Tian F, Li Y. Complete genomic characterization and identification of Saccharomycopsis phalluae sp. nov., a novel pathogen causes yellow rot disease on Phallus rubrovolvatus. J Fungi (Basel). 2021:7(9):707. doi: 10.3390/jof7090707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
New genome sequences reported in this study are listed in supplementary table S1, Supplementary Material online and have been deposited in the ENA/NCBI/DDBJ databases under BioProject PRJNA977123. Mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the following data set identifiers: PXD044638, PXD044720, PXD044729, PXD044731, PXD044788, PXD044742, PXD044763, PXD044772, PXD044785, PXD044786, PXD044693, and PXD044719.