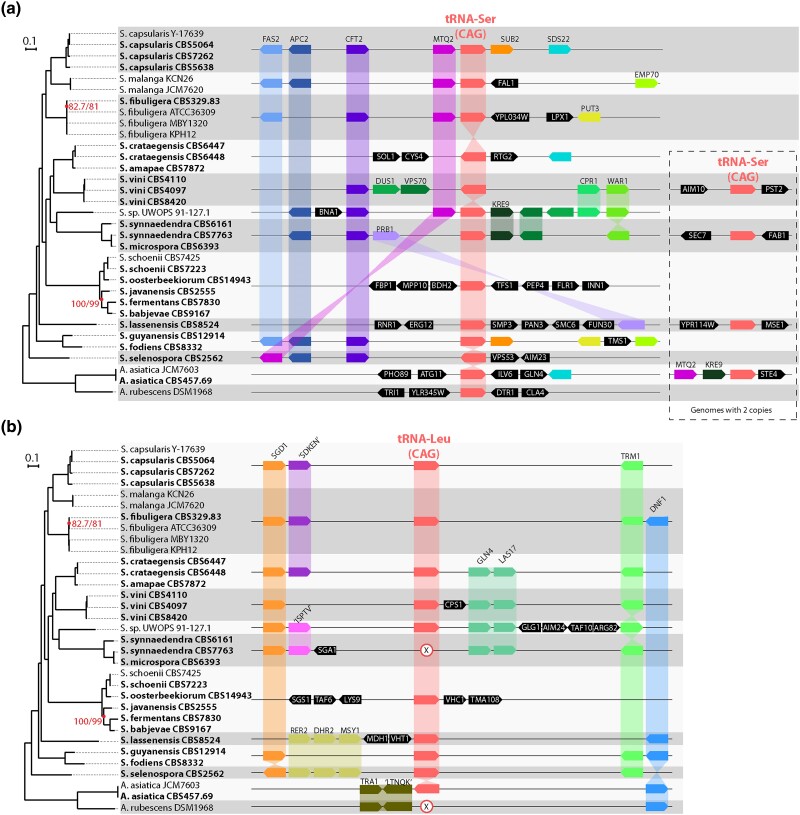
Fig. 1.
Phylogenomic tree of Saccharomycopsis and Ascoidea species, and synteny relationships around a) the tRNA-Ser(CAG) genes and b) the tRNA-Leu(CAG) genes. The same tree is shown in both panels. Strain names are indicated beside species names, and genomes sequenced in this study are indicated in bold. All branches except two were supported by 100% ultrafast bootstrapping and 100% SH-ALRT; the two branches not fully supported are indicated. a) Synteny relationships around tRNA-Ser(CAG) loci. Gray and white horizontal bands indicate groups of strains or species with identical gene arrangements, and only one gene diagram is shown for each group. Some genomes contain a second tRNA-Ser(CAG) gene, shown in the dashed box. In A. asiatica, one sequenced strain (JCM7603) contains two tRNA-Ser(CAG) genes but the other (CBS457.69) contains only one. The copy that is only in JCM7603 shares neighbors (MTQ2 and KRE9) with Saccharomycopsis species. b) Synteny relationships around the tRNA-Leu(CAG) locus. X symbols mark the absence of a tRNA-Leu(CAG) gene in genomes that have lost this gene. Genes are named according to their Saccharomyces cerevisiae orthologs where possible. Other genes are named by conserved sequence motifs they contain.