Graphical Abstract

Liver cancer is a complex disease following a chronic liver injury, inflammation, unregulated wound healing, fibrosis, or carcinogenesis.1 It also typically involves the activation of specific oncogenes and the inactivation of tumor suppressor genes.2 The oncoprotein gankyrin was first found to be overexpressed in hepatocellular carcinoma in 2000,3-7 while more recently its overexpression has also been associated with the proliferation and metastasis of numerous other cancers.1,4,8-13 The oncogenic nature of gankyrin is driven by its various protein-protein interactions (PPIs) (e.g., 26S proteasome, retinoblastoma protein (Rb),7,14,15 p53,7,14-17 cyclin-dependent kinase 4 (CDK4),14-18 and murine double minute 2 (MDM2)).14-17 Additionally, gankyrin promotes tumorigenicity by increasing Rb and p53 proteasomal degradation resulting in cell cycle proliferation.3,8,15,19,20 Interestingly, reduced gankyrin expression has been shown to decrease cell proliferation and colony formation in HepG2, HuH7, and Hep3B cells,1,18,21-23 while targeting gankyrin has shown promise in inhibiting tumor formation and metastasis.1,24 Due to its profound impact on the onset and development of various cancers through its numerous PPIs,11,22 gankyrin is an intriguing target for the treatment of certain cancers.
Despite being an attractive therapeutic target, there have been limited efforts in developing small molecule binders of gankyrin. In 2016, Chattopadhyay et al., discovered the first small molecule gankyrin binder (cjoc42, 1, Figure 1) that disrupts the gankyrin-proteasome interaction.25 Subsequent cell-based assays showed that cjoc42 treatment caused an increase in p53 levels, suggesting that cjoc42 disrupts the proteasomal degradation pathway.6,25 Cjoc42 alone and in combination with other chemotherapeutic agents has also shown an ability to decrease the proliferation of hepatoblastoma cells. This anti-proliferative effect was attributed to disruption of the gankyrin-26S proteasome interaction, resulting in increased levels of various tumor suppressor proteins (TSPs), such as Rb, p53, C/EBPα, HNF4α and CUGBP1.6,22,25 While substantial, the limited anti-proliferative activity (IC50 > 50 μM) points towards the need for further optimization of the cjoc42 scaffold to enhance its gankyrin-binding ability and anti-proliferative activity.
Figure 1.
Proposed binding of cjoc42 (Yellow) to Gankyrin (Blue) determined by molecular modeling and visualized using PyMol. PDB: 2DVW.26
Previous work from our group investigated substitutions to the phenyl rings, sulfonate ester, and triazole groups of cjoc42.12,27,28 Our goal for this project was to modify the propyl linker in combination with previously established alterations to cjoc42 to observe their impact on liver cancer proliferation and gankyrin binding (Figure 2).
Figure 2.
Chemical structures, thermal shift values, and anti-proliferative activity of cjoc42 (1), and proposed cjoc42 derivatives.
It has been previously suggested that cjoc42 binds to gankyrin primarily through interactions with Y15, R41, W46, S49, W74, and K116 (Figure 1).25 The focus of this study was to explore alterations to the three-carbon linker of the cjoc42 scaffold and determine their effect on gankyrin binding and anti-proliferative activity. Molecular modeling studies have suggested that cjoc42 adopts a U-shape conformation when bound to gankyrin. However, the propyl linker does not appear to directly interact with any particular amino acid residue of gankyrin (Figure 1). We hypothesized that adopting this U-shape is important for gankyrin binding to potentially maximize the π-π interactions with W46 and W74. To assess the importance of the flexibility of the propyl linker of cjoc42, the linker was substituted with a series of acyclic linkers. Derivatives were also designed to impart conformational constraint by reducing the distance between the two phenyl rings of cjoc42 by replacing the linker with either a urea or sulfonyl urea group and assessing their impact on anti-proliferative activity.
The synthesis of cjoc42 derivatives 4a-b (Scheme 1) commenced with a copper-catalyzed alkyne-azide cycloaddition (CuAAC) between methyl 4-azidobenzoate (2) and either 3-butyn-1-ol or 5-hexyn-1-ol, to afford triazole intermediates 3a-b. Triazole intermediates 3a-b then underwent nucleophilic substitution reactions with para-toluenesulfonyl chloride in the presence of triethylamine (TEA) and 4-dimethylaminopyridine (DMAP) to generate the desired cjoc42 derivatives 4a-b.
Scheme 1.
Synthesis of compounds 4a and 4b.a
a Reagents: (a) 3-Butyn-1-ol or 5-hexyn-1-ol, CuSO4, sodium ascorbate, THF/tBuOH/H2O (53-76%); (b) p-TsCl, DMAP, TEA, CH2Cl2, 0 °C (23-51%).
Cjoc42 derivatives 7a-c were then generated with the aim of replacing the sulfonate ester of cjoc42 with a previously optimized amide group12 in addition to replacing the propyl linker. The synthesis of derivatives 7a-c (Scheme 2) began with an EDCI-mediated amide coupling of para-toluic acid with various alkynyl amines to generate intermediates 6a-c. Amide intermediates 6a-c then underwent a CuAAC with methyl 4-azidobenzoate (2) to afford desired cjoc42 derivatives 7a-c.
Scheme 2.
Synthesis of compounds 7a-c.a
a Reagents: (a) Propargylamine, 3-butyn-1-amine, or 5-hexyn-1-amine, EDCI, pyridine, CH2Cl2 (47-76%) (b) Methyl 4-azidobenzoate, CuSO4, sodium ascorbate, THF/tBuOH/H2O (13-77%).
To evaluate the impact of these modifications, the anti-proliferative activity of compounds 7a-c was determined against the gankyrin overexpressing cell line, HuH6, as shown in Table 1. Replacing the carbon linker with either a 2 or 4 carbon linker (4a and 4b, respectively) did not improve the anti-proliferative activity as compared to cjoc42. However, compound 7c exhibited a modest improvement in anti-proliferative activity against HuH6 cells (IC50 = 19.5 μM). These findings were in agreement with our previous work where replacing the sulfonate ester of cjoc42 with an amide group improves anti-proliferative activity.29 These results also demonstrated that replacing the propyl linker with an ethyl or butyl linker did not significantly impact the anti-proliferative activity, suggesting that the linker length plays little to no role in influencing the anti-proliferative activity. Additionally, the improvement in anti-proliferative activity observed for 7c suggests that an amide group replacement has more impact on the activity than replacement of propyl linker alone.
Table 1.
| Compound | HuH6 IC50 (μM) |
|---|---|
| Cjoc42 | > 50 |
| 4a | > 50 |
| 4b | > 50 |
| 7a | > 50 |
| 7b | > 50 |
| 7c | 19.5 (± 2.5) |
HuH6 cells were incubated for 24 h prior to drug addition.
HuH6 cells were incubated for 72 h at 37 °C in 5% CO2 with the respective drug.
Cell proliferation was determined using an MTT assay.
All experiments were performed in replicates of 6-8.
Previous work from our group also showed that replacing the triazole group of cjoc42 with an amide group improved the scaffold’s anti-proliferative activity.12 Therefore, derivatives 10a-f were designed and synthesized with an aim of replacing the triazole and sulfonate ester groups of cjoc42 with amide groups while combining them with optimal alkyl linker lengths. The synthesis of derivatives 10a-f (Scheme 3) was performed with an EDCI-mediated amide coupling of either methyl 4-azidobenzoate (8a) or p-toluic acid (8b) with various diamines (9a-c) to generate cjoc42 derivatives 10a-f. Derivatives 13a-h then replaced the triazole and sulfonate ester groups with amide groups. Derivatives 13a-h (Scheme 4) were also synthesized by an EDCI-mediated amide coupling of either p-toluidine (11a) or methyl 4-aminobenzoate (11b) with various dicarboxylic acids (12a-d) to generate cjoc42 derivatives 13a-h.
Scheme 3.
Synthesis of compounds 10a-f.a
a Reagents: (a) EDCI, pyridine, CH2Cl2 (13-52%).
Scheme 4.
Synthesis of compounds 13a-h.a
a Reagents: (a) EDCI, pyridine and CH2Cl2 (6-46%).
Cjoc42 derivatives 10a-f and 13a-h were then evaluated for their anti-proliferative activity against HuH6 cells as shown in Table 2. Compounds 10a, 10d, and 13f showed modest improvements in anti-proliferative activity as compared to cjoc42. However, compound 13d substantially improved the anti-proliferative activity against HuH6 cells (IC50 = 3.3 μM). These results again demonstrated that replacement of the propyl linker with other alkyl linkers in combination with replacing the triazole and sulfonate ester groups of cjo42 with amide groups resulted in improved anti-proliferative activity.
Table 2.
| Compound | HuH6 IC50 (μM) |
|---|---|
| 10a | 30.2 (± 3.1) |
| 10b | > 50 |
| 10c | > 50 |
| 10d | 19.8 (± 2.5) |
| 10e | > 50 |
| 10f | > 50 |
| 13a | > 50 |
| 13b | > 50 |
| 13c | > 50 |
| 13d | 3.3 (±0.6) |
| 13e | >50 |
| 13f | 14.1 (± 1.5) |
| 13g | >50 |
| 13h | >50 |
HuH6 cells were incubated for 24 h prior to drug addition.
HuH6 cells were incubated for 72 h at 37 °C in 5% CO2 with the respective drug.
Cell proliferation was determined using an MTT assay.
All experiments were performed in replicates of 6-8.
Previous work from our lab demonstrated that a 4-phenyl substitution on the phenyl ring connected to the sulfonate ester of cjoc42 improved gankyrin binding and anti-proliferative activity. Additionally, a 4-trifluoromethyl or 4-methoxy substitution on the phenyl ring connected to the triazole of cjoc42 enhanced gankyrin binding and modestly improved its anti-proliferative activity against certain gankyrin overexpressing cancer cell lines.12,27,28 As previously discussed, compounds 10d, 13d, and 13f improved anti-proliferative activity against HuH6 cells, as compared to cjoc42. Therefore, cjoc42 derivatives 17a-j and 21a-b were designed to combine the optimized features of 10d, 13d, and 13f with the previously optimized aryl sulfonate ester and aryl triazole substitutions. The synthesis of derivatives 17a-j (Scheme 5) then began with an EDCI-mediated amide coupling between various p-substituted benzoic acid derivatives and specific mono-Boc-protected diamines 14a-c to generate intermediates 15a-e. These intermediates then underwent an acid-catalyzed decarboxylation to provide intermediates 16a-e. Lastly, intermediates 16a-e then underwent another EDCI-mediated amide coupling with specific p-substituted benzoic acids to generate desired derivatives 17a-j.
Scheme 5.
Synthesis of compounds 17a-j.a
a Reagents: (a) p-toluic acid, monomethyl terephthalate, [1,1′-biphenyl]-4-carboxylic acid, or 4-methoxybenzoic acid, EDCI, pyridine, and CH2Cl2 (47-74%); (b) 6 M HCl and EtOH, 0 °C (75-98%); (c) p-toluic acid, monomethyl terephthalate, 4-(trifluoromethyl) benzoic acid, [1,1′-biphenyl]-4-carboxylic acid, or 4-methoxybenzoic acid, EDCI, pyridine, and CH2Cl2 (13-48%).
The synthesis of compounds 21a-b (Scheme 6) began with an EDCI-mediated amide coupling of p-toluidine or 4-aminobiphenyl with monoesters 18a or 18b to generate intermediates 19a-b. Intermediates 19a-b then underwent ester hydrolysis to afford intermediates 20a-b followed by a second EDCI-mediated amide coupling with p-toluidine or methyl 4-aminobenzoate to generate desired cjoc42 derivatives 21a-b.
Scheme 6.
Synthesis of compounds 21a-b.a
a Reagents: (a) p-toluidine or 4-aminobiphenyl, EDCI, pyridine, CH2Cl2 (75-80%); (b) 10 M NaOH and MeOH (90-98%); (c) p-toluidine or methyl 4-aminobenzoate, EDCI, pyridine, CH2Cl2 (15-37%).
Derivatives 17a-j and 21a-b were then evaluated for their anti-proliferative activity against HuH6 cells as shown in Table 3. Compounds 17a and 17c showed modest improvements in anti-proliferative activity while compound 17e (IC50 = 2.4 μM) showed a substantial improvement in activity against HuH6 cells, as compared to cjoc42. This further demonstrated that a 4-phenyl substitution or 4-methoxy substitution improves the anti-proliferative activity against HuH6 cells. However, changing the propyl linker length resulted in little to no impact on the anti-proliferative activity.
Table 3.
| Compound | HuH6 IC50 (μM) |
|---|---|
| 17a | 31.0 (± 5.0) |
| 17b | > 50 |
| 17c | 20.3 (± 1.4) |
| 17d | > 50 |
| 17e | 2.4 (± 0.2) |
| 17f | > 50 |
| 17g | > 50 |
| 17h | > 50 |
| 17i | > 50 |
| 17j | > 50 |
| 21a | > 50 |
| 21b | > 50 |
HuH6 cells were incubated for 24 h prior to drug addition.
HuH6 cells were incubated for 72 h at 37 °C in 5% CO2 with the respective drug.
Cell proliferation was determined using an MTT assay.
All experiments were performed in replicates of 6-8.
Compounds 23a-b, 24a-c, 26a-b, and 27a-c were then designed and synthesized to incorporate a constrained linker (urea and sulfonyl urea: Schemes 7 and 8) in place of the propyl linker to assess its impact on anti-proliferative activity. The syntheses of compounds 23a-e and 25a-e were performed with a nucleophilic addition between either p-tolyl isocyanate or p-toluenesulfonyl isocyanate with various anilines or benzylamines to generate desired compounds 23a-e and 25a-e.
Scheme 7.
Synthesis of compounds 23a-e.a
a Reagents: p-toluidine, methyl 4-aminobenzoate, 4-methoxybenzylamine, or (aminomethyl) benzoate, DIPEA, CH2Cl2 (97-99%).
Scheme 8.
Synthesis of compounds 25a-e.a
a Reagents: p-toluidine, methyl 4-aminobenzoate, 4-methoxybenzylamine, or methyl 4-(aminomethyl) benzoate, DIPEA, CH2Cl2 (95-100%).
Compounds 23a-e and 25a-e were then evaluated for their anti-proliferative activity against HuH6 cells, as shown in Table 4. Interestingly, only modest to no improvement in anti-proliferative activity was observed, suggesting that reducing the distance and flexibility of the linker between the phenyl rings of the cjoc42 scaffold has a detrimental effect on anti-proliferative activity.
Table 4.
| Compound | HuH6 (μM) |
|---|---|
| 23a | > 50 |
| 23b | 38.8 (± 1.9) |
| 23c | > 50 |
| 23d | > 50 |
| 23e | > 50 |
| 25a | > 50 |
| 25b | > 50 |
| 25c | > 50 |
| 25d | > 50 |
| 25e | > 50 |
HuH6 cells were incubated for 24 h prior to drug addition.
HuH6 cells were incubated for 72 h at 37 °C in 5% CO2 with the respective drug.
Cell proliferation was determined using an MTT assay.
All experiments were performed in replicates of 6-8.
Anti-proliferative activity against non-cancerous cells
To assess the relative selectivity of these next-generation cjoc42 derivatives, the most potent compounds were also evaluated against the non-cancerous HEK-293 cell line (Table 5). Compounds 10d and 13d exhibited IC50 values > 50 μM against HEK-293, demonstrating they preferentially target cancer cells and potentially target gankyrin which is overexpressed in HuH6 cells but not HEK-293 cells. Interestingly, compound 17e (IC50 =3.3 μM) showed similar potency against both HEK-293 cells and HuH6 cells, suggesting that this compound exhibits little to no selectivity for cancer cells and therefore was not pursued further. Compound 13d was then chosen for subsequent biological evaluation due to its potent anti-proliferative activity and high degree of selectivity for cancer cells.
Table 5.
| Compound | Structure | HuH6 IC50 (μM) | HEK-293 IC50(μM) |
|---|---|---|---|
| 10d |

|
19.8 (± 2.5) | > 50 |
| 13d |

|
3.3 (± 0.6) | > 50 |
| 17e |

|
2.4 (± 0.2) | 3.3 (± 0.3) |
HuH6 and HEK293 cells were incubated for 24 h prior to drug addition.
HuH6 and HEK-293 cells were incubated for 72 h at 37 °C in 5% CO2 with the respective drug.
Cell proliferation was determined using an MTT assay.
All experiments were performed in replicates of 6-8.
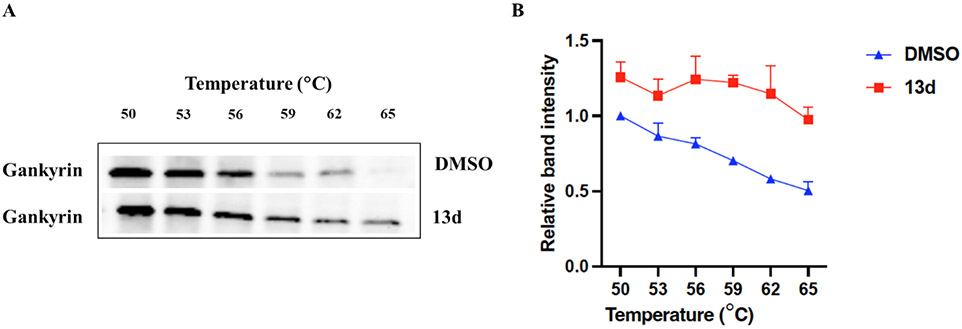
Cellular Thermal Shift Assay (CETSA)
To determine if compound 13d effectively binds gankyrin in HuH6 cells, it was evaluated in a cellular thermal shift assay (CETSA, Figure 3). When HuH6 cells were treated with compound 13d, it demonstrated the ability to bind and thermally stabilize gankyrin in HuH6 cells at its respective anti-proliferative IC50 value (Figure 3A). Interestingly, this is the first gankyrin-binding small-molecule to demonstrate a stabilizing effect on gankyrin by CETSA and these results suggest that compound 13d effectively binds gankyrin in HuH6 cells.
Figure 3.
CETSA to assess the binding of 13d to gankyrin. (A) Representative blot of gankyrin stability in HuH6 cells treated with compound 13d for 5 h. (B) CETSA melt curve for gankyrin from HuH6 cells in the presence of compound 13d. Data represents the mean ± SD (n = 3).
Western Blot Analysis
Gankyrin facilitates the proteasomal degradation of essential TSPs, such as p53 and Rb, and previous work with cjoc42 and subsequent derivatives has demonstrated an ability to bind gankyrin and increase the levels of p53 and/or Rb in various cancers.6,25 Western blot analysis of compound 13d in HuH6 cells was then performed (Figure 4B). Our data shows that 13d caused an increase in both Rb and p53 levels, while gankyrin levels were relatively constant, which is in agreement with previous data for cjoc42 and some of its derivatives.12,22 This suggests that compound 13d exhibits a similar mechanism of action to cjoc42 and its derivatives by disrupting the proteasomal degradation pathway resulting in reduced cancer cell proliferation.
Figure 4.
Western blot analysis of HuH6 cells treated with compound 13d. (A) Chemical structure and IC50 value of 13d in HuH6 cells. (B) Western blot analysis of TSP levels from HuH6 lysate upon treatment with 13d. All experiments were conducted in duplicate, and a representative result is shown.
Cell Cycle Analysis
Gankyrin’s critical role in regulating p53 and Rb levels in turn results in cell cycle regulation.4,8 As previously discussed, compound 13d significantly improved the anti-proliferative activity against HuH6 cells while also increasing levels of Rb and p53, therefore it was selected for cell cycle analysis in HuH6 cells (Figure 5). Rb and p53 are essential TSPs for cell cycle regulation, where p53 upregulation is known to arrest the cell cycle in the G1 phase,30,31 and Rb is a checkpoint for G1 while restricting S-phase entry.31,32 Treatment of HuH6 cells with 3.3 μM of compound 13d caused a significant increase in the number of cells in the G0-G1 phase and a significant decrease in the number of cells in the S phase (Figure 5). This suggests that compound 13d halts cell cycle progression in the G0-G1 phase, which is further supported by our western blot results (Figure 4). This data also suggests that the possible mode of action of 13d is gankyrin binding resulting in disruption of the proteasomal degradation pathway and ultimately inhibition of cell cycle progression.
Figure 5.
Cell cycle analysis of compound 13d in HuH6 cells. (A) Cell cycle analysis by flow cytometry of HuH6 cells treated with DMSO. (B) Cell cycle analysis by flow cytometry of HuH6 cells treated with compound 13d. (C) Representative histogram data of the cell cycle analysis of HuH6 cells treated with DMSO and compound 13d. Data represents the mean ± SD (n = 3). Note: *0.01 < p < 0.05.
Anti-proliferative activity against other liver cancer cell lines
Since compound 13d significantly improved anti-proliferative activity against HuH6 cells, it was also evaluated for its anti-proliferative activity against three other gankyrin-overexpressing liver cancer cell lines: HuH7, HepG2, and Hep3B (Table 6). Compound 13d then showed a modest improvement in anti-proliferative activity against HepG2 cells with an IC50 of 24.0 μM. However, no improvement in activity was observed against Hep3B and HuH7 cells (IC50 > 50 μM). The lack of potency against Hep3B and HuH7 cells could be due to their p53 status, as Hep3B is p53-null33 and HuH7 has a mutant p53 status,34 while HuH6 and HepG2 possess a wild-type p53 status.35 Additionally, HuH6 and HepG2 cells are a type of hepatoblastoma (HBL),35 while HuH7 and Hep3B are hepatocellular carcinoma cells (HCC). 34,35 These results suggest that both p53 status and liver cancer type (HBL vs. HCC) play important roles in the anti-proliferative activity of cjoc42 derivatives.
Table 6.
| Cell line | IC50 (μM) |
|---|---|
| HepG2 | 24.0 (± 2.6) |
| Hep3B | >50 |
| HuH7 | > 50 |
HuH7, HepG2 and Hep3B cells were incubated for 24 h prior to drug addition. HuH7, HepG2 and Hep3B cells were incubated for 72 h at 37 °C in 5% CO2 with the respective drug.
Cell proliferation was determined using an MTT assay.
All experiments were performed in replicates of 6-8.
Compound effects on gankyrin conformation
Circular Dichroism (CD) was performed to assess the effect of compounds lOd and 13d on gankyrin’s global 3D structural fold. CD is a robust biophysical method that can be used to monitor the α-helical content of gankyrin over a gradient of increasing compound concentration. To assess the specificity of compounds 10d and 13d towards gankyrin, we also conducted parallel analyses on ANKRA2, another ankyrin-repeating containing protein with a similar 3D fold to gankyrin.
The CD spectra for gankyrin and ANKRA2 alone revealed characteristic spectra minima at 208 nm and 222 nm confirming α-helical content of the native state. Subsequent examination of gankyrin spectra in the presence of each small molecule revealed a significant loss of gankyrin structural integrity, indicating that compounds 10d and 13d destabilize gankyrin’s 3D fold. Conversely, the spectra of ANKRA2 in the presence of both small molecules exhibits no significant global structure change. These findings confirm that compounds 10d and 13d are specific for targeting gankyrin and disrupt its structural integrity, with no effect on ANKRA2.
Conclusion
To assess the importance of the flexibility of the cjoc42 propyl linker on its gankyrin binding ability and anti-proliferative activity, various acyclic linkers were evaluated in combination with triazole and sulfonate ester replacements. Compound 13d was found to significantly improve anti-proliferative activity against HuH6 cells, while CETSA experiments showed that compound 13d effectively binds gankyrin in HuH6 cells. Our western blot analyses also showed that compound 13d could effectively increase levels of p53 and Rb in HuH6 cells, demonstrating its ability to bind gankyrin and inhibit the proteasomal degradation pathway. This is also supported by our CD spectra that shows compound 13d disrupts the global fold of gankyrin which is a core component of the 19S regulatory cap of the 26S proteasome. Compound 13d was then able to arrest the cell cycle of HuH6 cells in the G0-G1 phase, which was expected due to the observed increases in Rb and p53 levels. Our cumulative data suggests that replacing the propyl linker with alkyl linkers of various lengths did not have a major impact on the potency of the derivatives. However, replacing the propyl linker of cjoc42 along with appropriate triazole and sulfonate ester isosteres improved gankyrin specific binding and anti-proliferative activity. Compound 13d also modestly improved anti-proliferative against another gankyrin overexpressing cell line, HepG2. Interestingly, no improvement in activity was observed for Hep3B and HuH7, which do over express gankyrin but are either have p53 null or mutated p53 status in addition to being HCC cell lines.
In summary, these results indicate that compound 13d is only effective against cell lines with wild-type p53 status (HuH6 and HepG2) and are HBL cell lines. This finding indicates a potentially unique mechanism by which these cjoc42 derivatives inhibit the gankyrin-proteasome interaction. Although further evaluation is required to fully understand the exact mechanism of these derivatives, they can be used to further clarify gankyrin’s role in HBL development.
Materials and Methods
All commercially available reagents were purchased and used without further purification. Commercially available solvents (> 99.0% purity) were used for column chromatography without further purification. Proton (Ή) and carbon (13C) NMR spectra were recorded on a 400 MHz spectrometer. Proton chemical shifts are reported in ppm (δ) relative to residual Acetone-d6 (2.05 ppm), DMSO-d6 (2.49 ppm), CDCl3 (7.26 ppm), or Methanol-d4 (4.78 ppm, 3.31) ppm. Data are reported as follows: chemical shift (multiplicity [singlet (s), doublet (d), doublet of doublets (dd), doublet of doublets of doublets (ddd), triplet (t), quartet (q), quintet (p), sextet (h), multiplet (m)], coupling constants [Hz], integration). Carbon chemical shifts are reported in ppm with the respective solvent resonance as the internal standard: Acetone-d6 (206.7 ppm and 29.9 ppm), DMSO-d6 (39.5 ppm), CDCl3 (77.2 ppm), or Methanol-d4 (49.15 ppm). Unless otherwise noted, all NMR spectra were acquired at ambient temperature. Analytical thin-layer chromatography (TLC) was performed using Silica Gel 60 Å F254 precoated plates (0.25 mm thickness). UV absorption was used as the primary visualization method for monitoring reactions and column chromatography. UPLC/HPLC was performed using reverse-phase C18 column with UV detector. Accurate mass measurements/high resolution mass spectra (HRMS) were obtained from the Columbia University Chemistry Department Mass Spectrometry Facility on a Waters Xevo G2-XS QToF mass spectrometer equipped with an H-Class UPLC inlet and a LockSpray ESI source.
Purity analysis
Purity analysis of compounds 4a-b, 7a-c, 10a-f, 13a-h, 17a-j, 21a-b, 23a-e, and 25a-e was carried out using an Agilent 1260 infinity series HPLC system (Agilent, Santa Clara, CA). Purity analysis was performed using an Agilent Eclipse plus C18, 3.5 μm, 4.6 mm × 100 mm column and the runs were monitored at 254 nm. Methanol and water mixtures were used as the mobile phase. An isocratic run was performed with 90% methanol in water over 5 min, and the flow rate was 0.5 mL/min. Purity analysis of compounds 4a-b, 7a-c, 10a-f, 13a-h, 17a-j, 21a-b, 23a-e, and 25a-e was carried out using an Agilent 1100 infinity series HPLC system (Agilent, Santa Clara, CA). Purity analysis was performed using an Agilent Eclipse plus C18, 3.5 μm, 4.6 mm × 100 mm column and the runs were monitored at 254 nm. Methanol and water mixtures were used as the mobile phase. An isocratic run was performed with 90% methanol in water over 12 min with a flow rate of 0.5 mL/min.
All target compounds were analyzed to be > 95% pure by the methods described above unless otherwise noted.
General procedure 1 - Amide coupling.
To a stirred solution of carboxylic acid (1 equiv) in dichloromethane was added EDCI (3 equiv) and amine (1 equiv), followed by the addition of pyridine (5 equiv). The reaction mixture was then stirred overnight at room temperature. The reaction mixture was then diluted with dichloromethane and washed 3 times with water. The organic layer was then dried over sodium sulfate, filtered, concentrated under reduced pressure, and purified by column chromatography, unless otherwise stated.
General procedure 2 - Copper-catalyzed azide-alkyne cycloaddition.
To a stirred solution of azide (1 equiv) in a THF/tBuOH/H2O (2:3:5) mixture was added alkyne (1 equiv), copper sulfate (0.3 M, 0.3 equiv), and sodium ascorbate (0.1 M, 0.01 equiv). The reaction mixture was then stirred overnight at room temperature. Upon completion, the reaction mixture was partitioned between ethyl acetate and water, and then washed 3 times with water. The organic layer was then dried over sodium sulfate, filtered, concentrated under reduced pressure, and purified by column chromatography, unless otherwise stated.
General procedure 3 – Urea and sulfonylurea formation.
To a stirred solution of either isocyanate or sulfonyl isocyanate (1 equiv) in dichloromethane was added amine (1 equiv) and DIPEA (3 equiv). The reaction mixture was then stirred overnight at room temperature. Upon completion, the reaction mixture was partitioned between ethyl acetate and water, and then washed 3 times with water. The organic layer was then dried over sodium sulfate, filtered, concentrated under reduced pressure, and purified by column chromatography, unless otherwise stated.
General procedure 4 - Tosylation.
To a stirred solution of an alcohol intermediate in dichloromethane at 0 °C was added triethylamine (1 equiv) and 4-dimethylaminopyridine (0.1 equiv). p-Toluene sulfonyl chloride (1.5 equiv) was then added, and the reaction mixture was stirred for an additional 15 min at 0 °C. The reaction mixture was then stirred overnight at room temperature. Upon completion, the reaction mixture was partitioned between dichloromethane and water. The organic layer was then washed 3 times with water, dried over sodium sulfate, filtered, and the solvent was removed under reduced pressure. The compounds were then purified using column chromatography.
General procedure 5 – Boc deprotection.
To a stirred solution of Boc-protected intermediates 16a, 16b, 16c, 16d or 16e in ethanol at 0 °C was added 6M HCl, and the reaction mixture was stirred for an additional 1 h at 0 °C. The reaction mixture was then stirred overnight at room temperature. Upon completion, the reaction mixture was concentrated to obtain the desired compounds which were then used in the next reaction without further purification.
General procedure 6 – Ester hydrolysis.
Ester intermediates 19a or 19b were dissolved in MeOH, followed by addition of 10M NaOH, and the reaction mixture was then stirred overnight at room temperature. Upon completion, the reaction mixture was concentrated to remove MeOH. The crude mixture was then partitioned between dichloromethane and water, and the organic layer was then washed 3 times with water, dried over sodium sulfate, filtered, and the solvent was removed under reduced pressure to obtain the desired carboxylic acid intermediate which was used in the next reaction without further purification.
Methyl 4-(4-(2-hydroxyethyl)-1H-1,2,3-triazol-1-yl) benzoate (3a)
General procedure 2, (150 mg, 53%). 1HNMR (400 MHz, DMSO-d6) δ 8.70 (s, 1H), 8.19 – 8.09 (m, 2H), 8.10 – 7.97 (m, 2H), 4.82 (s, 1H), 3.88 (s, 3H), 3.72 (d, J = 3.1 Hz, 2H), 2.87 (t, J = 6.8 Hz, 2H). 13C NMR (100 MHz, DMSO-d6) δ 165.81, 146.56, 140.38, 131.42 (2C), 129.43, 121.30, 119.95 (2C), 60.57, 52.82, 29.59.
Methyl 4-(4-(4-hydroxybutyl)-1H-1,2,3-triazol-1-yl) benzoate (3b)
General procedure 2, (248 mg, 76%). 1H NMR (400 MHz, CDCl3) δ 8.20 (d, J = 8.8 Hz, 2H), 7.85 (d, J = 8.8 Hz, 2H), 7.85 (s, 1H), 3.98 (d, J = 6.3 Hz, 3H), 3.73 (t, J = 6.4 Hz, 2H), 2.87 (t, J = 7.5 Hz, 2H), 1.87 (dt, J = 12.6, 7.4 Hz, 3H), 1.72 (dd, J = 14.9, 6.5 Hz, 2H). 13C NMR (100 MHz, CDCl3) δ 165.98, 149.22, 140.18, 131.30, 129.87, 119.66 (3C), 118.88, 62.22, 32.06, 25.49 (2C), 25.23.
Methyl 4-(4-(2-(tosyloxy)ethyl)-1H-1,2,3-triazol-1-yl)benzoate (4a)
General procedure 4, (75 mg, 23%). 1H NMR (400 MHz, CDCl3) δ 8.30 – 8.17 (m, 2H), 7.91 (s, 1H), 7.90 – 7.72 (m, 4H), 7.32 (d, J = 8.0 Hz, 2H), 4.38 (t, J = 6.3 Hz, 2H), 3.99 (s, 3H), 3.21 (t, J = 6.2 Hz, 2H), 2.43 (s, 3H). 13C NMR (100 MHz, CDCl3) δ 165.90, 148.44, 144.84, 140.18, 132.98, 131.27 (2C), 129.89 (2C), 127.81 (2C), 119.64 (2C), 118.98 (2C), 70.23, 52.38, 28.15, 25.11, 24.79, 21.60. HRMS (ESI+): m/z calculated for C19H19N3O5S+ [M+H]+ 402.1045, found 402.1124.
Methyl 4-(4-(4-(tosyloxy)butyl)-1H-1,2,3-triazol-1-yl)benzoate (4b)
General procedure 4, (198 mg, 51%). 1H NMR (400 MHz, CDCl3) δ 8.13 (d, J = 8.7 Hz, 2H), 7.76 (d, J = 9.5 Hz, 3H), 7.71 (d, J = 8.3 Hz, 2H), 7.27 (d, J = 8.2 Hz, 2H), 4.00 (t, J = 5.8 Hz, 2H), 3.89 (s, 3H), 2.72 (t, J = 7.0 Hz, 2H), 2.37 (s, 3H), 1.77 – 1.67 (m, 4H). 13C NMR (100 MHz, CDCl3) δ 165.90, 148.44, 144.84, 140.18, 132.98, 131.27 (2C), 129.89 (2C), 127.81 (2C), 119.64 (2C), 118.98 (2C), 70.23, 52.38, 28.22, 25.11, 24.79, 21.60. HRMS (ESI+): m/z calculated for C21H23N3O5S+ [M+H]+ 430.1358, found 430.1452.
4-methyl-N-(prop-2-yn-1-yl)benzamide (6a)
General procedure 1, (160 mg, 47%). 1H NMR (400 MHz, CDCl3) δ 7.71 (d, J = 8.2 Hz, 2H), 7.25 (d, J = 7.9 Hz, 2H), 6.39 (s, 1H, NH), 4.26 (dd, J = 5.2, 2.6 Hz, 2H), 2.41 (s, 3H), 2.29 (t, J = 2.5 Hz, 1H). 13C NMR (100 MHz, CDCl3) δ 167.15, 142.25, 130.91, 129.26 (2C), 127.09 (2C), 79.69, 71.73, 29.71, 21.47.
N-(but-3-yn-1-yl)-4-methylbenzamide (6b)
General procedure 1, (330 mg, 56%). 1H NMR (400 MHz, DMSO-d6) δ 8.58 (s, 1H), 7.74 (d, J = 8.0 Hz, 2H), 7.27 (d, J = 7.9 Hz, 2H), 2.85 (s, 1H, NH), 2.51 (s, 2H), 2.41 (d, J = 2.4 Hz, 2H), 2.35 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 166.56, 141.53, 131.96, 129.27 (2C), 127.62 (2C), 82.73, 72.57, 38.80, 21.40, 19.17.
N-(hex-5-yn-1-yl)-4-methylbenzamide (6c)
General procedure 1, (110 mg, 76%). 1H NMR (400 MHz, CDCl3) δ 7.68 (d, J = 8.1 Hz, 2H), 7.15 (d, J = 7.9 Hz, 2H), 6.94 (s, 1H, NH), 3.39 (dd, J = 13.0, 6.7 Hz, 2H), 2.35 (s, 3H), 2.17 (td, J = 6.9, 2.4 Hz, 2H), 1.95 (t, J = 2.5 Hz, 1H), 1.80 – 1.59 (m, 2H), 1.61 – 1.47 (m, 2H). 13C NMR (100 MHz, CDCl3) δ 167.74, 141.59, 131.84, 129.07 (2C), 127.02 (2C), 84.12, 68.76, 39.45, 28.70, 25.80, 21.39, 18.10.
Methyl 4-(4-((4-methylbenzamido)methyl)-1H-1,2,3-triazol-1-yl)benzoate (7a)
General procedure 2, (80 mg, 30%). 1H NMR (400 MHz, CDCl3) δ 8.22 – 8.06 (m, 3H), 7.76 (d, J = 8.5 Hz, 2H), 7.65 (d, J = 8.0 Hz, 2H), 7.21 – 7.13 (m, 2H), 7.11 (s, 1H, NH), 4.72 (d, J = 5.7 Hz, 2H), 3.88 (s, 3H),2.31 (s, 3H). 13C NMR (100 MHz, CDCl3) δ 167.58, 165.89, 146.05, 142.29, 140.01, 131.34 (2C), 130.97, 130.29, 129.29 (2C), 127.05 (2C), 120.87, 119.92 (2C), 52.47, 35.30, 21.47. HRMS (ESI+): m/z calculated for C19H18N4O3+ [M+H]+ 351.1379 found 351.1476.
Methyl 4-(4-(2-(4-methylbenzamido)ethyl)-1H-1,2,3-triazol-1-yl)benzoate (7b)
General procedure 2, purified by recrystallization, (25 mg, 12.85%). 1H NMR (400 MHz, DMSO-d6) δ 8.80 (s, 1H), 8.58 (s, 1H, NH), 8.16 (d, J = 8.2 Hz, 2H), 8.07 (d, J = 8.4 Hz, 2H), 7.74 (d, J = 7.7 Hz, 2H), 7.26 (d, J = 7.8 Hz, 2H), 3.90 (s, 3H), 3.59 (d, J = 6.0 Hz, 2H), 2.99 (t, J = 7.0 Hz, 2H), 2.34 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 166.65, 165.84, 146.59, 141.44, 140.40, 132.17, 131.49 (2C), 129.55, 129.25 (2C), 127.65 (2C), 121.31, 120.07 (2C), 52.87, 26.01, 21.40 (2C). HRMS (ESI+): m/z calculated for C20H20N4O3+ [M+H]+ 365.1535, found 365.1617.
Methyl 4-(4-(4-(4-methylbenzamido)butyl)-1H-1,2,3-triazol-1-yl)benzoate (7c)
General procedure 2, (140 mg, 77%). 1H NMR (400 MHz, CDCl3) δ 8.19 (d, J = 8.8 Hz, 2H), 7.89 (s, 1H), 7.84 (d, J = 8.8 Hz, 2H), 7.69 (d, J = 8.1 Hz, 2H), 7.22 (d, J = 7.9 Hz, 2H), 6.52 (s, 1H, NH), 3.97 (s, 3H), 3.51 (dd, J = 12.9, 6.8 Hz, 2H), 2.87 (t, J = 7.3 Hz, 2H), 2.39 (s, 3H), 1.93 – 1.81 (m, 2H), 1.81 – 1.67 (m, 2H). 13C NMR (100 MHz, CDCl3) δ 167.60, 165.97, 148.82, 141.79, 140.20, 131.73, 131.30 (2C), 129.93, 129.20 (2C), 126.88 (2C), 119.70 (2C), 119.02, 52.43, 39.64, 28.94, 26.50, 25.00, 21.42. HRMS (ESI+): m/z calculated for C22H24N4O3+ [M+H]+ 393.1848, found 393.1927.
N,N’-(ethane-1,2-diyl)bis(4-methylbenzamide) (10a)
General procedure 1, purified by trituration, (244 mg, 22%). 1H NMR (400 MHz, DMSO-d6) δ 8.53 (s, 2H), 7.76 (d, J = 8.0 Hz, 4H), 7.26 (d, J = 7.9 Hz, 4H), 3.49 – 3.38 (m, 4H), 2.35 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 166.96 (2C), 141.41 (2C), 132.21 (2C), 129.49 (4C), 127.67 (4C), 40.21 (2C), 21.40 (2C). HRMS (ESI+): m/z calculated for C18H20N2NaO2+ [M+Na]+ 319.1422 found 319.1437. This matches the previously reported spectra.36
Dimethyl 4,4'-((ethane-1,2-diylbis(azanediyl))bis(carbonyl))dibenzoate (10b)
General procedure 1, purified by trituration, (120 mg, 9%). 1H NMR (400 MHz, DMSO-d6) δ 8.83 (s, 2H), 8.03 (s, 4H), 7.98 (s, 4H), 3.88 (s, 6H), 3.47 (d, J = 2.6 Hz, 4H). 13C NMR (100 MHz, DMSO-d6) δ 166.21 (2C) δ 166.15 (2C), 139.13 (2C), 132.17 (2C), 129.58 (4C), 128.10 (4C), 52.84 (2C), 40.78 (2C). HRMS (ESI+): m/z calculated for C20H21N2O6+ [M+H]+ 385.1321, found 385.1423. This matches the previously reported spectra.36.
N,N'-(propane-1,3-diyl)bis(4-methylbenzamide) (10c)
General procedure 1, (490 mg, 52%). 1H NMR (400 MHz, DMSO-d6) δ 8.44 (t, J = 5.4 Hz, 2H), 7.75 (d, J = 8.1 Hz, 4H), 7.27 (d, J = 7.9 Hz, 4H), 3.33 – 3.23 (m, 4H), 2.35 (s, 6H), 1.84 – 1.67 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 165.78 (2C), 146.97, 140.00, 131.63 (2C), 131.07, 130.97 (2C), 128.89 (2C), 122.09, 120.49 (2C), 52.95 (2C), 39.51 (2C), 21.69. HRMS (ESI+): m/z calculated for C19H22N2NaO2+ [M+Na]+ 333.1579, found 333.1580.
Dimethyl 4,4'-((propane-1,3-diylbis(azanediyl))bis(carbonyl))dibenzoate (10d)
General procedure 1, (450 mg, 38%). 1H NMR (400 MHz, DMSO-d6) δ 8.72 (t, J = 5.4 Hz, 2H), 8.04 (d, J = 8.4 Hz, 4H), 7.96 (d, J = 8.4 Hz, 4H), 3.88 (s, 6H), 3.33 (s, 4H), 1.88 – 1.74 (m, 2H). 13C NMR (100 MHz, CDCl3) δ 167.29, 166.31, 138.08 (2C), 132.85 (2C), 129.94 (4C), 127.07 (4C), 77.22 (2C), 52.42, 36.33, 29.80. HRMS (ESI+): m/z calculated for C21H23N2O6+ [M+H]+ 399.1556, found 399.1562. This matches the previously reported spectra.36
N,N'-(butane-1,4-diyl)bis(4-methylbenzamide) (10e)
General procedure 1, purified by trituration, (130 mg, 16%). 1H NMR (400 MHz, DMSO-d6) δ 8.40 (t, J = 5.2 Hz, 2H), 7.74 (d, J = 7.9 Hz, 4H), 7.25 (d, J = 7.9 Hz, 4H), 3.27 (d, J = 5.1 Hz, 4H), 2.34 (s, 6H), 1.55 (s, 4H). 13C NMR (100 MHz, DMSO-d6) δ 166.52 (2C), 141.43 (2C), 132.41 (2C), 129.20 (4C), 127.61 (4C), 40.19 (2C), 27.26 (2C), 21.44 (2C). HRMS (ESI+): m/z calculated for C20H24N2NaO2+ [M+Na]+ 347.1735, found 347.1825.
Dimethyl 4,4'-((butane-1,4-diylbis(azanediyl))bis(carbonyl))dibenzoate (10f)
General procedure 1, purified by trituration, (120 mg, 13%), 1H NMR (400 MHz, DMSO-d6) δ 8.69 (t, J = 5.1 Hz, 2H), 8.03 (d, J = 8.2 Hz, 4H), 7.95 (d, J = 8.2 Hz, 4H), 3.88 (s, 6H), 3.31 (d, J = 5.7 Hz, 4H), 1.59 (s, 4H). 13C NMR (100 MHz, DMSO-d6) δ 166.22 (2C), 165.77 (2C), 139.23 (2C), 132.09 (2C), 129.57 (4C), 128.01 (4C), 52.82 (2C), 39.35 (2C), 27.04 (2C). HRMS (ESI+): m/z calculated for C22H25N2O6+ [M+H]+ 413.1713, found 413.1713. This matches the previously reported spectra.36
N1,N3-di-p-tolylmalonamule (13a)
General procedure 1, (277 mg, 30%). 1H NMR (400 MHz, DMSO-d6) δ 10.05 (s, 2H, NH), 7.48 (d, J = 8.4 Hz, 4H), 7.11 (d, J = 8.2 Hz, 4H), 3.42 (s, 2H), 2.25 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 165.69 (2C), 136.94 (2C), 132.75 (2C), 129.60 (4C), 119.56 (4C), 46.27, 20.91 (2C). HRMS (ESI+): m/z calculated for C17H18N2NaO2+ [M+Na]+ 305.1266, found 305.1282. This matches the previously reported spectra.27
Dimethyl 4,4'-(malonylbis(azanediyl)) dibenzoate (13b)
General procedure 1, (120 mg, 31%). 1H NMR (400 MHz, DMSO-d6) δ 10.56 (s, 2H, NH), 7.94 (d, J = 8.7 Hz, 4H), 7.75 (d, J = 8.7 Hz, 4H), 3.83 (s, 6H), 3.58 (s, 2H). 13C NMR (100 MHz, DMSO-d6) δ 166.39 (4C), 143.64 (2C), 130.82 (4C), 124.64 (2C), 118.98 (4C), 52.37 (2C), 46.70. HRMS (ESI+): m/z calculated for C19H19N2O6+ [M+H]+ 371.1243, found 371.1258.
N1,N4-di-p-tolylsuccinamide (13c)
General procedure 1, purified by recrystallization, (110 mg, 13%). 1H NMR (400 MHz, DMSO-d6) δ 9.87 (s, 2H), 7.46 (d, J = 8.2 Hz, 4H), 7.08 (d, J = 8.1 Hz, 4H), 2.62 (s, 4H), 2.23 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 170.59 (2C), 137.42 (2C), 132.21(2C), 129.49 (4C), 119.68 (4C), 31.68 (2C), 20.88 (2C). HRMS (ESI+): m/z calculated for C18H20N2NaO2+ [M+Na]+ 319.1422, found 319.1434. This matches the previously reported spectra.37
Dimethyl 4,4'-(succinylbis(azanediyl))dibenzoate (13d)
Purified by recrystallization. General procedure 1, (50 mg, 7%). 1H NMR (400 MHz, DMSO-d6) δ 10.38 (s, 2H), 7.91 (d, J = 8.7 Hz, 4H), 7.73 (d, J = 8.7 Hz, 4H), 3.82 (s, 6H), 2.74 (s, 4H). 13C NMR (100 MHz, DMSO-d6) δ 171.61 (2C), 166.53 (2C), 144.14 (2C), 130.57 (4C), 124.05 (2C), 118.62 (4C), 52.48 (2C), 31.74 (2C). HRMS (ESI+): m/z calculated for C20H19N2NaO6+ [M+Na]+ 407.1219, found 407.1222.
N1,N5-di-p-tolylglutaramide (13e)
General procedure 1, (121 mg, 46%). 1H NMR (400 MHz, DMSO-d6) δ 9.82 (s, 2H), 7.48 (d, J = 8.2 Hz, 4H), 7.09 (d, J = 7.8 Hz, 4H), 2.35 (t, J = 7.3 Hz, 4H), 2.24 (s, 6H), 1.96 – 1.81 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 171.02 (2C), 137.30 (2C), 132.25 (2C), 129.47 (4C), 119.43 (4C), 36.00 (2C), 21.44, 20.89 (2C). HRMS (ESI+): m/z calculated for C19H22N2Nao2+ [M+Na]+ 333.1579, found 333.1587.
Dimethyl 4,4'-(glutaroylbis(azanediyl))dibenzoate (13f)
General procedure 1, (60 mg, 10%). 1H NMR (400 MHz, DMSO) δ 10.31 (s, 2H), 7.91 (d, J = 7.5 Hz, 4H), 7.75 (d, J = 7.7 Hz, 4H), 3.82 (s, 6H), 2.44 (t, J = 7.2 Hz, 4H), 2.25 (t, J = 7.7 Hz, 2H). 13C NMR (100 MHz, CDCl3) δ 167.60, 165.97, 148.82, 141.79, 140.20, 131.73, 131.30 (2C), 129.93, 129.20 (2C), 126.88 (2C), 119.70 (2C), 119.02, 52.43, 39.64, 28.94, 26.50, 25.00, 21.42. HRMS (ESI+): m/z calculated for C21H22N2O6+ [M+H]+ 399.1566, found 399.1566.
N1,N6-di-p-tolyladipamide (13g)
General procedure 1, (97 mg, 10%). 1H NMR (400 MHz, DMSO-d6) δ 9.79 (s, 2H), 7.46 (d, J = 8.2 Hz, 4H), 7.08 (d, J = 8.2 Hz, 4H), 2.30 (s, 4H), 2.23 (s, 6H), 1.61 (s, 4H). 13C NMR (100 MHz, DMSO-d6) δ 171.32 (2C), 137.26 (2C), 132.28 (2C), 129.47 (4C), 119.55 (4C), 36.70 (2C), 25.40 (2C), 20.88 (2C). HRMS (ESI+): m/z calculated for C20H25N2O2+ [M+H]+ 325.1916, found 325.1919. This matches the previously reported spectra.38
Dimethyl 4,4’-(adipoylbis(azanediyl))dibenzoate (13h)
General procedure 1, (36 mg, 6%). 1H NMR (400 MHz, DMSO-d6) δ 10.26 (s, 2H), 7.90 (d, J = 7.8 Hz, 4H), 7.73 7.73 (d, J = 7.8 Hz, 4H), 3.84 (d, J = 16.8 Hz, 6H), 2.39 (s, 4H), 1.65 (s, 4H). 13C NMR (100 MHz, DMSO-d6) δ 172.18 (2C), 166.30 (2C), 144.16 (2C), 130.73 (4C), 124.13 (2C), 118.82 (4C), 52.32 (2C), 36.81 (2C), 25.12 (2C). HRMS (ESI+): m/z calculated for C22H25N2O6+ [M+H]+ 413.1713, found 413.1715.39,40
tert-butyl (2-(4-methylbenzamido)ethyl)carbamate (15a)
General procedure 1, (255 mg, 47%). 1H NMR (400 MHz, CDCl3) δ 7.71 (d, J = 8.0 Hz, 2H), 7.21 (d, J = 8.1 Hz, 2H), 7.16 (s, 1H), 5.09 (s, 1H), 3.54 (dd, J = 10.8, 5.2 Hz, 2H), 3.40 (d, J = 5.1 Hz, 2H), 2.38 (s, 3H), 1.43 (s, 9H). 13C NMR (100 MHz, CDCl3) δ 167.85, 157.46, 141.79, 131.32, 129.13 (2C), 127.01 (2C), 79.90, 41.90, 40.03, 28.34 (3C), 21.43.
tert-butyl (3-(4-methylbenzamido)propyl)carbamate (15b)
General procedure 1, (300 mg, 59%). 1H NMR (400 MHz, CDCl3) δ 7.71 (d, J = 7.6 Hz, 2H), 7.60 (s, 1H), 7.15 (d, J = 7.7 Hz, 2H), 5.43 (s, 1H), 3.44 (dd, J = 11.8, 5.9 Hz, 2H), 3.16 (d, J = 4.8 Hz, 2H), 2.33 (s, 3H), 1.64 (s, 2H), 1.40 (s, 9H). 13C NMR (100 MHz, CDCl3) δ 167.78, 156.90, 141.58, 131.67, 129.06 (2C), 127.05 (2C), 79.17, 37.08, 36.21, 30.06, 28.39 (3C), 21.37.
tert-butyl (3-([1,1'-biphenyl]-4-carboxamido)propyl)carbamate (15c)
General procedure 1, (410 mg, 73%). 1H NMR (400 MHz, DMSO-d6) δ 8.51 (t, J = 5.4 Hz, 1H), 7.94 (d, J = 8.3 Hz, 2H), 7.75 (dd, J = 15.7, 7.9 Hz, 4H), 7.50 (t, J = 7.5 Hz, 2H), 7.42 (d, J = 7.3 Hz, 1H), 6.85 (s, 1H), 3.28 (dd, J = 12.8, 6.6 Hz, 2H), 2.99 (dd, J = 12.7, 6.4 Hz, 2H), 1.74 – 1.59 (m, 2H), 1.38 (s, 9H). 13C NMR (100 MHz, CDCl3) δ 167.21, 157.03, 144.10, 140.16, 133.24, 128.89 (2C), 127.90, 127.53, 127.21 (2C), 79.60, 36.04, 30.29, 28.41 (3C).
tert-butyl (4-(4-methylbenzamido)butyl)carbamate (15d)
General procedure 1, (350 mg, 74%). 1H NMR (400 MHz, CDCl3) δ 7.69 (d, J = 7.9 Hz, 2H), 7.20 (d, J = 7.9 Hz, 2H), 6.63 (s, 1H), 4.74 (s, 1H), 3.46 (q, J = 6.5 Hz, 2H), 3.15 (d, J = 6.2 Hz, 2H), 2.38 (s, 3H), 1.60 (ddd, J = 21.4, 10.7, 5.1 Hz, 4H), 1.44 (s, 9H). 13C NMR (100 MHz, CDCl3) δ 167.57, 156.21, 141.70, 131.76, 129.13 (2C), 126.96 (2C), 79.24, 40.38, 39.58, 28.42 (3C), 27.69, 26.69, 21.42.
tert-butyl (4-([1,1'-biphenyl]-4-carboxamido)butyl)carbamate (15e)
General procedure 1, (250 mg, 64%). 1H NMR (400 MHz, CDCl3) δ 7.81 (d, J = 7.9 Hz, 2H), 7.56 (dd, J = 15.2, 7.8 Hz, 4H), 7.39 (t, J = 7.5 Hz, 2H), 7.32 (t, J = 7.3 Hz, 1H), 6.49 (s, 1H), 4.59 (s, 1H), 3.45 (dd, J = 12.5, 6.4 Hz, 2H), 3.11 (d, J = 5.9 Hz, 2H), 1.62 – 1.51 (m, 4H), 1.38 (s, 9H). 13C NMR (100 MHz, CDCl3) δ 167.28, 156.23, 144.14, 140.09, 133.31, 128.90, 127.94, 127.50, 127.19 (2C), 127.16, 79.28, 39.67, 28.42 (3C), 27.78, 26.48.
N-(2-aminoethyl)-4-methylbenzamide (16a)
General procedure 5, (250 mg, 98%). 1H NMR (400 MHz, DMSO-d6) δ 8.82 (t, J = 4.7 Hz, 1H), 8.30 (s, 2H), 7.87 (d, J = 7.9 Hz, 2H), 7.29 (t, J = 10.5 Hz, 2H), 3.56 (s, 2H), 3.01 (d, J = 5.4 Hz, 2H), 2.35 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 167.09, 141.70, 131.62, 129.20 (2C), 127.93 (2C), 39.04, 37.53, 21.42.
N-(3-aminopropyl)-4-methylbenzamide (16b)
General procedure 5, (255 mg, 85%). 1H NMR (400 MHz, DMSO) δ 8.74 (d, J = 4.9 Hz, 1H), 8.15 (s, 2H), 7.81 (d, J = 7.8 Hz, 2H), 7.27 (d, J = 7.8 Hz, 2H), 3.33 (dd, J = 11.8, 5.9 Hz, 2H), 2.82 (d, J = 5.7 Hz, 2H), 2.32 (d, J = 21.7 Hz, 3H), 1.91 – 1.74 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 166.75, 141.51, 131.92, 129.25 (2C), 127.71 (2C), 37.10, 36.53, 27.70, 21.41.
N-(3-aminopropyl)-[1,1'-biphenyl/-4-carboxamide (16c)
General procedure 5, (645 mg, 81%). 1H NMR (400 MHz, DMSO-d6) δ 8.86 (s, 1H), 8.09 (s, 2H), 8.00 (d, J = 8.2 Hz, 2H), 7.76 (dd, J = 17.6, 7.8 Hz, 4H), 7.50 (t, J = 7.6 Hz, 2H), 7.42 (d, J = 7.2 Hz, 1H), 3.37 (dd, J = 12.3, 6.2 Hz, 2H), 2.85 (dd, J = 12.7, 6.4 Hz, 2H), 1.89 – 1.81 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 166.54, 143.22, 139.62, 133.51, 129.51 (2C), 128.53, 128.39 (2C), 127.33 (2C), 126.96 (2C), 37.16, 36.66, 27.74.
N-(4-aminobutyl)-4-methylbenzamide (16d)
General procedure 5, (200 mg, 90%). 1H NMR (400 MHz, DMSO-d6) δ 8.55 (t, J = 5.1 Hz, 1H), 8.07 (s, 2H), 7.78 (d, J = 7.9 Hz, 2H), 7.26 (d, J = 7.9 Hz, 2H), 3.34 – 3.17 (m, 2H), 2.79 (d, J = 5.5 Hz, 2H), 2.35 (s, 3H), 1.58 (s, 4H). 13C NMR (100 MHz, DMSO-d6) δ 166.51, 141.32, 132.23, 129.20 (2C), 127.67 (2C), 38.95, 38.84, 26.63, 24.95, 21.40.
N-(4-aminobutyl)-[1,1'-biphenyl]-4-carboxamide (16e)
General procedure 5, (188 mg, 75%). 1H NMR (400 MHz, DMSO-d6) δ 8.64 (t, J = 5.5 Hz, 1H), 7.96 (d, J = 8.3 Hz, 2H), 7.75 (dd, J = 16.5, 7.8 Hz, 4H), 7.56 – 7.36 (m, 3H), 3.31 (d, J = 5.5 Hz, 2H), 2.81 (d, J = 5.5 Hz, 2H), 1.60 (s, 4H). 3C NMR (100 MHz, DMSO-d6) δ 166.29, 143.10, 139.65, 133.81, 129.50, 128.51, 128.32, 127.32 (2C), 126.93, 39.05, 38.92, 26.63, 25.02.
Methyl 4-((2-(4-methylbenzamido)ethyl)carbamoyl)benzoate (17a)
General procedure 1, (240 mg, 46%). 1H NMR (400 MHz, DMSO-d6) δ 8.82 (s, 1H), 8.55 (s, 1H), 8.04 (d, J = 8.3 Hz, 2H), 7.97 (d, J = 8.3 Hz, 2H), 7.76 (d, J = 8.0 Hz, 2H), 7.26 (d, J = 8.0 Hz, 2H), 3.88 (s, 3H), 3.45 (s, 4H), 2.35 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 166.87, 166.21, 166.13, 141.42, 139.14, 132.20, 132.17, 129.58 (2C), 129.23 (2C), 128.09 (2C), 127.68 (2C), 52.83 (2C), 21.40 (2C). HRMS (ESI+): m/z calculated for C19H21N2O4+ [M+H]+ 341.1501, found 341.1517.
Methyl 4-((3-(4-methylbenzamido)propyl)carbamoyl)benzoate (17b)
General procedure 1, (200 mg, 48%). 1H NMR (400 MHz, CDCl3) δ 8.11 (d, J = 8.0 Hz, 2H), 7.98 (d, J = 8.1 Hz, 2H), 7.76 (d, J = 7.9 Hz, 2H), 7.72 (d, J = 5.8 Hz, 1H), 7.27 (t, J = 7.5 Hz, 2H), 7.08 (s, 1H), 3.96 (s, 3H), 3.56 (dd, J = 12.8, 6.4 Hz, 4H), 2.42 (s, 3H), 1.84 (d, J = 4.9 Hz, 2H). 13C NMR (100 MHz, CDCl3) δ 168.49, 166.99, 166.41, 142.20, 138.29, 132.64, 131.20, 129.84 (2C), 129.34 (2C), 127.15 (2C), 126.99 (2C), 52.37 (2C), 36.32, 29.86, 21.46. HRMS (ESI+): m/z calculated for C20H23N2O4+ [M+H]+ 355.1658, found 355.1686.
N-(3-(4-methylbenzamido)propyl)-[1,1'-biphenyl]-4-carboxamide (17c)
General procedure 1, purified by trituration, (200 mg, 16%). 1H NMR (400 MHz, DMSO-d6) δ 8.70 (t, J = 5.5 Hz, 1H), 8.57 (t, J = 5.5 Hz, 1H), 8.01 (d, J = 8.4 Hz, 2H), 7.83 (d, J = 8.2 Hz, 4H), 7.81 – 7.76 (m, 2H), 7.55 (t, J = 7.5 Hz, 2H), 7.46 (t, J = 7.3 Hz, 1H), 7.32 (d, J = 8.0 Hz, 2H), 3.41 – 3.36 (m, 4H), 2.40 (s, 3H), 1.85 (p, J = 6.8 Hz, 2H). 13C NMR (100 MHz, DMSO-d6) δ 166.59, 166.34, 143.08, 141.34, 139.68, 133.87, 132.28, 129.49 (2C), 129.25 (2C), 128.48, 128.29 (2C), 127.61 (2C), 127.32 (2C), 126.96 (2C), 37.44, 29.78, 21.40 (2C). HRMS (ESI+): m/z calculated for C24H25N2O2+ [M+H]+ 373.1916, found 373.1932.
Methyl 4-((3-([1,1'-biphenyl]-4-carboxamido)propyl)carbamoyl)benzoate (17d)
General procedure 1, (45 mg, 13%). 1H NMR (400 MHz, DMSO-d6) δ 8.77 (s, 1H), 8.61 (s, 1H), 8.13 – 7.85 (m, 6H), 7.75 (dd, J = 14.4, 7.8 Hz, 4H), 7.47 (dd, J = 24.9, 17.4 Hz, 3H), 3.88 (s, 3H), 3.30 – 3.23 (m, 2H), 1.88 – 1.76 (m, 2H), 1.23 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 166.35, 166.21, 165.88, 143.10, 139.68, 139.18, 133.85, 132.15, 129.61 (2C), 129.49 (2C), 128.49, 128.27 (2C), 128.01 (2C), 127.33 (2C), 126.96 (2C), 52.83, 37.75, 37.60, 29.62. HRMS (ESI+): m/z calculated for C25H25N204+ [M+H]+ 417.1814, found 417.1840.
N-(3-(4-methoxybenzamido)propyl)-[1,1'-biphenyl]-4-carboxamide (17e)
General procedure 1, purified by trituration, (72 mg, 15%). 1H NMR (400 MHz, DMSO-d6) δ 8.58 (s, 1H), 8.39 (s, 1H), 7.94 (d, J = 8.2 Hz, 2H), 7.83 (d, J = 8.6 Hz, 2H), 7.75 (dd, J = 16.5, 7.8 Hz, 4H), 7.50 (t, J = 7.5 Hz, 2H), 7.42 (d, J = 7.2 Hz, 1H), 7.00 (d, J = 8.6 Hz, 2H), 3.80 (s, 3H), 3.31 (s, 4H), 1.86 – 1.70 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 166.34, 166.22, 161.93, 143.09, 139.67, 133.86, 129.49 (2C), 129.39 (2C), 128.49, 128.27 (2C), 127.33 (2C), 127.28, 126.97 (2C), 113.93 (2C), 55.79, 37.57, 37.45, 29.88. HRMS (ESI+): m/z calculated for C24H25N2NaO3+ [M+Na]+ 411.1685, found 411.1698.
N-(3-(4-(trifluoromethyl)benzamido)propyl)-[1,1'-biphenyl]-4-carboxamide (17f)
General procedure 1, purified by trituration, (69 mg, 20%). 1H NMR (400 MHz, DMSO-d6) δ 8.77 (d, J = 5.2 Hz, 1H), 8.59 (t, J = 5.2 Hz, 1H), 8.05 (d, J = 8.0 Hz, 2H), 7.95 (d, J = 8.1 Hz, 2H), 7.86 (d, J = 8.1 Hz, 2H), 7.75 (dd, J = 16.2, 7.9 Hz, 4H), 7.50 (t, J = 7.5 Hz, 2H), 7.42 (d, J = 7.2 Hz, 1H), 3.35 – 3.29 (m, 2H), 2.51 (s, 2H), 1.93 – 1.73 (m, 2H). 13C NMR (100 MHz, DMSO-d6) δ 166.37, 165.54, 143.11, 139.67, 138.82, 133.84, 131.65, 131.34, 129.50 (2C), 128.52 (2C), 128.28 (2C), 127.33 (2C), 126.97 (2C), 125.75, 123.09, 40.90, 37.77, 37.58, 29.61. HRMS (ESI+): m/z calculated for C24H22F3N2O2+ [M+H]+ 427.1633, found 427.1638.
Methyl 4-((4-(4-methylbenzamido)butyl)carbamoyl)benzoate (17g)
General procedure 1, (150 mg, 27%). 1H NMR (400 MHz, DMSO-d6) δ 8.68 (s, 1H), 8.39 (s, 1H), 8.03 (d, J = 8.3 Hz, 2H), 7.96 (d, J = 7.8 Hz, 2H), 7.74 (d, J = 7.7 Hz, 2H), 7.25 (d, J = 7.9 Hz, 2H), 3.88 (s, 3H), 3.30 (dd, J = 9.4, 4.3 Hz, 4H), 2.35 (s, 3H), 1.57 (s, 4H). 13C NMR (100 MHz, DMSO-d6) δ 166.44, 166.21, 165.74, 141.26, 139.25, 132.34, 132.08, 129.57 (2C), 129.20 (2C), 128.02 (2C), 127.61 (2C), 52.82, 27.23 (2C), 27.08 (2C), 21.39. HRMS (ESI+): m/z calculated for C21H25N2O4+ [M+H]+ 369.1814, found 369.1833.
Methyl 4-((4-([1,1'-biphenyl]-4-carboxamido)butyl)carbamoyl)benzoate (17h)
General procedure 1, purified by trituration, (150 mg, 30%). 1H NMR (400 MHz, DMSO-d6) δ 8.70 (s, 1H), 8.54 (s, 1H), 8.10 – 8.01 (m, 4H), 7.99 – 7.91 (m, 3H), 7.74 (dd, J = 12.9, 7.9 Hz, 3H), 7.48 (d, J = 7.6 Hz, 2H), 7.41 (t, J = 7.1 Hz, 1H), 3.88 (d, J = 4.6 Hz, 4H), 3.32 – 3.28 (m, 3H), 1.59 (s, 4H). 13C NMR (100 MHz, DMSO-d6) δ 166.22, 165.76, 143.02, 139.69, 139.26, 133.93, 132.08, 129.57 (2C), 129.49 (2C), 128.47, 128.29 (2C), 128.02 (2C), 127.32 (2C), 126.92 (2C), 52.82, 27.19 (2C), 27.08 (2C). HRMS (ESI+): m/z calculated for C26H26N2NaO4+ [M+Na]+ 453.1790, found 453.1805.
N-(4-(4-methylbenzamido)butyl)-[1,1'-biphenyl]-4-carboxamide (17i)
General procedure 1, (203 mg, 45%). 1H NMR (400 MHz, DMSO-d6) δ 8.54 (t, J = 5.4 Hz, 1H), 8.40 (t, J = 5.3 Hz, 1H), 7.94 (d, J = 8.3 Hz, 2H), 7.82 – 7.67 (m, 6H), 7.49 (s, 2H), 7.41 (t, J = 7.3 Hz, 1H), 7.25 (d, J = 7.9 Hz, 2H), 3.30 (dd, J = 10.5, 5.5 Hz, 4H), 2.34 (s, 3H), 1.58 (s, 4H). 13C NMR (100 MHz, DMSO-d6) δ 166.45, 166.21, 143.02, 141.26, 139.69, 133.94, 132.35, 129.49 (2C), 129.20 (2C), 128.47, 128.29 (2C), 127.61 (2C), 127.32 (2C), 126.92 (2C), 39.47, 27.23 (2C), 21.39 (2C). HRMS (ESI+): m/z calculated for C25H27N2O2+ [M+H]+ 387.2073, found 387.2090.
N-(4-(4-(trifluoromethyl)benzamido)butyl)-[1,1'-biphenyl]-4-carboxamide (17j)
General procedure 1, purified by trituration, (120 mg, 23%). 1H NMR (400 MHz, DMSO-d6) δ 8.74 (t, J = 5.3 Hz, 1H), 8.55 (t, J = 5.4 Hz, 1H), 8.05 (d, J = 8.1 Hz, 2H), 7.95 (d, J = 8.3 Hz, 2H), 7.85 (d, J = 8.2 Hz, 2H), 7.75 (dd, J = 14.0, 7.8 Hz, 4H), 7.50 (t, J = 7.5 Hz, 2H), 7.41 (t, J = 7.3 Hz, 1H), 3.33 (d, J = 3.4 Hz, 4H), 1.61 (s, 4H). 13C NMR (100 MHz, DMSO-d6) δ 166.23, 165.43, 143.03, 139.69, 138.89, 133.93, 131.53, 129.48 (2C), 128.53 (2C), 128.47, 128.29 (2C), 127.31 (2C), 126.92 (2C), 125.77, 125.73, 27.19 (2C), 27.05 (2C). HRMS (ESI+): m/z calculated for C25H23F3N2NaO2+ [M+Na]+ 463.1609, found 463.1619.
Methyl 4-([1,1'-biphenyl]-4-ylamino)-4-oxobutanoate (19a)
General procedure 1, (1975 mg, 75%). 1H NMR (400 MHz, CDCl3) δ 7.92 (s, 1H), 7.54 (d, J = 13.9 Hz, 6H), 7.40 (s, 2H), 7.35 – 7.15 (m, 1H), 3.71 (s, 3H), 2.73 (d, J = 25.4 Hz, 4H). 13C NMR (100 MHz, CDCl3) δ 173.76, 169.89, 140.50, 137.23, 137.07, 128.79 (2C), 127.56 (2C), 127.09, 126.83 (2C), 120.16 (2C), 52.05, 32.05, 29.26.
Methyl 6-oxo-6-(p-tolylamino)hexanoate (19b)
General procedure 1, (286 mg, 80%). 1H NMR (400 MHz, CDCl3) δ 7.73 (s, 1H), 7.42 (d, J = 8.0 Hz, 2H), 7.11 (d, J = 8.0 Hz, 2H), 3.68 (s, 3H), 2.36 (t, J = 6.8 Hz, 4H), 2.31 (s, 3H), 1.81 – 1.63 (m, 4H). 13C NMR (100 MHz, CDCl3) δ 174.08, 170.91, 135.46, 133.77, 129.40 (2C), 120.03 (2C), 51.61, 37.06, 33.68, 25.00, 24.33, 20.85.
4-([1,1'-biphenyl]-4-ylamino)-4-oxobutanoic acid (20a)
General procedure 6, (1530 mg, 90%). 1H NMR (400 MHz, DMSO-d6) δ 11.80 (s, 1H), 7.69 – 7.51 (m, 6H), 7.43 (t, J = 7.3 Hz, 2H), 7.31 (t, J = 7.3 Hz, 1H), 2.45 – 2.33 (m, 2H), 2.23 (t, J = 6.4 Hz, 2H). 13C NMR (100 MHz, DMSO-d6) δ 176.24, 175.14, 172.95, 140.33, 139.92, 134.47, 129.32 (2C), 127.25 (2C), 126.61 (2C), 119.53 (2C), 35.30, 34.59.
6-oxo-6-(p-tolylamino)hexanoic acid (20b)
General procedure 6, (263 mg, 92%). 1H NMR (400 MHz, DMSO-d6) δ 12.11 (s, 1H), 9.85 (s, 1H), 7.53 (d, J = 8.2 Hz, 2H), 7.15 (d, J = 8.2 Hz, 2H), 2.43 – 2.30 (m, 4H), 2.28 (s, 3H), 1.76 – 1.52 (m, 4H). 13C NMR (100 MHz, DMSO-d6) δ 174.85, 171.23, 137.28, 132.25, 129.48 (2C), 119.51 (2C), 36.52, 33.89, 25.15, 24.62, 20.89.
Methyl 4-(4-([1,1'-biphenyl]-4-ylamino)-4-oxobutanamido)benzoate (21a)
General procedure 1, purified by trituration, (40 mg, 13%). 1H NMR (400 MHz, DMSO-d6) δ 10.09 (s, 1H), 9.90 (s, 1H), 7.70 (s, 1H), 7.62 (t, J = 9.0 Hz, 3H), 7.54 – 7.40 (m, 4H), 7.32 (t, J = 7.3 Hz, 1H), 7.09 (d, J = 8.2 Hz, 2H), 2.50 (s, 4H), 2.24 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 170.95, 170.54, 140.20, 139.29, 137.31, 135.02, 132.20, 129.51 (2C), 129.49, 129.35 (2C), 127.42, 127.34 (2C), 126.66 (2C), 119.74, 119.45, 119.39, 31.78, 31.64, 20.89. HRMS (ESI+): m/z calculated for C23H22N2NaO2+ [M+Na]+ 381.1579 found 381.1578.
Methyl 4-(6-oxo-6-(p-tolylamino)hexanamido)benzoate (21b)
General procedure 1, (130 mg, 37%). 1H NMR (400 MHz, DMSO-d6) δ 10.26 (s, 1H), 9.80 (s, 1H), 7.97 – 7.88 (m, 2H), 7.74 (t, J = 9.8 Hz, 2H), 7.52 – 7.38 (m, 2H), 7.08 (d, J = 8.1 Hz, 2H), 3.82 (s, 3H), 2.35 (d, J = 27.5 Hz, 4H), 2.24 (s, 3H), 1.63 (s, 4H). 13C NMR (100 MHz, DMSO-d6) δ 172.25, 171.30, 166.32, 144.14, 137.24, 132.30, 130.72 (2C), 129.48 (2C), 124.14, 119.55 (2C), 118.84 (2C), 52.33, 36.84, 36.66, 25.33, 25.19, 20.88. HRMS (ESI+): m/z calculated for C21H24N2NaO4+ [M+Na]+ 391.1634, found 391.1649.
1,3-di-p-tolylurea (23a)
General procedure 3, purified by trituration, (320 mg, 89%). 1H NMR (400 MHz, DMSO-d6) δ 8.50 (s, 2H), 7.33 (d, J = 8.3 Hz, 4H), 7.08 (d, J = 8.2 Hz, 4H), 2.24 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 153.01, 137.59 (2C), 131.02 (2C), 129.62 (4C), 118.67 (4C), 20.80 (2C). HRMS (ESI+): m/z calculated for C15H17N2O+ [M+H]+ 241.1341, found 241.1361. This matches the previously reported spectra.41-43.
Methyl 4-(3-(p-tolyl)ureido)benzoate (23b)
General procedure 3, purified by trituration, (100 mg, 24%). 1H NMR (400 MHz, DMSO-d6) δ 9.06 (s, 1H), 8.70 (s, 1H), 7.89 (d, J = 8.7 Hz, 2H), 7.58 (d, J = 8.6 Hz, 2H), 7.35 (d, J = 8.3 Hz, 2H), 7.10 (d, J = 8.2 Hz, 2H), 3.82 (s, 3H), 2.25 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 166.17, 152.74, 144.97, 137.17, 131.56, 130.88 (2C), 129.70 (2C), 122.76, 118.98 (2C), 117.69 (2C), 52.24, 20.82. HRMS (ESI+): m/z calculated for C16H17N2O3+ [M+H]+ 285.1239, found 285.1252.
1-(4-methylbenzyl)-3-(p-tolyl)urea (23c)
General procedure 3, purified by trituration, (450 mg, 71%) 1H NMR (400 MHz, DMSO-d6) δ 8.39 (s, 1H), 7.27 (d, J = 8.3 Hz, 2H), 7.15 (dd, J = 19.5, 7.9 Hz, 4H), 7.02 (d, J = 8.2 Hz, 2H), 6.48 (t, J = 5.8 Hz, 1H), 4.23 (d, J = 5.8 Hz, 2H), 2.27 (s, 3H), 2.21 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 155.73, 138.38, 137.80, 136.19, 130.23, 129.50 (2C), 129.30 (2C), 127.60 (2C), 118.24 (2C), 42.95, 21.13, 20.76. HRMS (ESI+): m/z calculated for C16H19N2O+ [M+H]+ 255.1497, found 255.1521.
Methyl 4-((3-(p-tolyl)ureido)methyl)benzoate(23d)
General procedure 3, purified by trituration, (180 mg, 36%). 1H NMR (400 MHz, DMSO-d6)) δ 8.52 (s, 1H), 7.93 (d, J = 8.1 Hz, 2H), 7.43 (d, J = 8.0 Hz, 2H), 7.28 (d, J = 8.3 Hz, 2H), 7.03 (d, J = 8.2 Hz, 2H), 6.66 (s, 1H), 4.36 (d, J = 5.9 Hz, 2H), 3.84 (s, 3H), 2.21 (s, 3H). 13C NMR (100 MHz, DMSO-d6 δ 166.61, 155.79, 146.84, 138.28, 130.36, 129.71 (2C), 129.51 (2C), 128.48, 127.65 (2C), 118.34 (2C), 52.51, 42.96, 20.76. HRMS (ESI+): m/z calculated for C17H19N2O3+ [M+H]+ 299.1396, found 299.1412.
1-(4-methoxybenz.yl)-3-(p-tolyl)urea (23e)
General procedure 3, purified by trituration, (145 mg, 36%). 1H NMR (400 MHz, DMSO-d6)) δ 8.38 (s, 1H), 7.27 (d, J = 8.2 Hz, 2H), 7.22 (d, J = 8.3 Hz, 2H), 7.02 (d, J = 8.2 Hz, 2H), 6.89 (ds, J = 8.3 Hz, 2H), 6.46 (d, J = 5.4 Hz, 1H), 4.20 (d, J = 5.8 Hz, 2H), 3.73 (s, 3H), 2.21 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 158.64, 155.69, 138.38, 132.76, 130.20, 129.50 (2C), 128.97 (2C), 118.22 (2C), 114.17 (3C), 55.52, 42.67, 20.76. HRMS (ESI+): m/z calculated for C16H18N2NaO2+ [M+Na]+ 293.1266, found 293.1284.
4-methyl-N-(p-tolylcarbamoyl)benzenesulfonamide (25a)
General procedure 3, purified by recrystallization, (205 mg, 67%). 1H NMR (400 MHz, CDCl3) δ 9.95 (s, 1H), 8.05 (s, 1H), 7.90 (d, J = 8.2 Hz, 2H), 7.28 (d, J = 8.1 Hz, 2H), 7.21 (d, J = 8.3 Hz, 2H), 7.03 (d, J = 8.2 Hz, 2H), 2.39 (s, 3H), 2.26 (s, J = 8.4 Hz, 3H). 13C NMR (100 MHz, CDCl3) δ 149.45, 144.37, 136.95, 134.87, 133.35, 129.57 (2C), 129.39 (2C), 127.68 (2C), 119.65 (2C), 21.59, 20.76. HRMS (ESI+): m/z calculated for C15H17N2O3S+ [M+H]+ 305.0960 found 305.1461. This matches the previously reported spectra.44,45
Methyl 4-(3-tosylureido)benzoate (25b)
General procedure 3, purified by trituration, (140 mg, 30%). 1H NMR (400 MHz, DMSO-d6) δ 10.96 (s, 1H), 9.25 (s, 1H), 7.86 (d, J = 8.3 Hz, 4H), 7.46 (dd, J = 15.6, 8.4 Hz, 4H), 3.80 (s, 3H), 2.40 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 166.81, 166.20, 149.81, 144.43, 143.15, 137.38, 131.53, 130.80, 129.98, 129.76, 128.03, 126.08, 118.64, 113.11, 52.35, 51.60. HRMS (ESI+): m/z calculated for C16H16N2NaO5S+ [M+Na]+ 371.0678, found 371.0694. This matches the previously reported spectra.45,46
4-methyl-N-((4-methylbenzyl)carbamoyl)benzenesulfonamide (25c)
General procedure 3, purified by trituration, (100 mg, 24%). 1H NMR (400 MHz, DMSO-d6) δ 7.58 (d, J = 8.2 Hz, 2H), 7.27 (d, J = 8.0 Hz, 2H), 7.22 (d, J = 8.0 Hz, 2H), 7.17 (d, J = 7.9 Hz, 2H), 6.33 (s, 2H), 2.34 (s, 3H), 2.30 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 154.99, 141.51, 141.24, 137.05, 129.37 (3C), 128.89 (2C), 128.43 (2C), 127.38 (2C), 21.38 (2C), 21.17 HRMS (ESI+): m/z calculated for C16H19N2NaO3S+ [M+Na]+ 341.0936 found 341.0948.
Methyl 4-((3-tosylureido)methyl)benzoate (25d)
General procedure 3, purified by trituration, (150 mg, 45%). 1H NMR (400 MHz, DMSO-d6) δ 10.89 (s, 1H), 7.92 (d, J = 8.2 Hz, 2H), 7.85 (d, J = 8.2 Hz, 2H), 7.47 (d, J = 8.1 Hz, 2H), 7.30 (d, J = 8.1 Hz, 2H), 7.19 (t, J = 5.7 Hz, 1H), 4.29 (d, J = 5.9 Hz, 2H), 3.90 (s, 3H), 2.46 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 166.52, 152.11, 145.42, 144.14, 137.77, 129.93, 129.62 (2C), 128.61, 127.71 (2C), 127.55 (2C), 52.53, 42.98, 21.48. HRMS (ESI+): m/z calculated for C17H18N2NaO5S+ [M+Na]+ 385.0834, found 385.0834.
N-((4-methoxybenzyl)carbamoyl)-4-methylbenzenesulfonamide (25e)
General procedure 3, purified by trituration, (90 mg, 18%). 1H NMR (400 MHz, DMSO-d6) δ 10.64 (s, 1H), 7.78 (d, J = 8.1 Hz, 2H), 7.41 (d, J = 8.1 Hz, 2H), 7.07 (d, J = 8.4 Hz, 2H), 6.91 (s, 1H), 6.83 (d, J 8.3 Hz, 2H), 4.07 (d, J = 5.7 Hz, 2H), 3.72 (s, 3H), 2.40 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 158.83, 151.86, 144.08, 137.84, 131.51, 129.91 (2C), 128.93 (2C), 127.71 (2C), 114.12 (2C), 55.50, 42.64, 21.50. HRMS (ESI+): m/z calculated for C16H18N2NaO4S+ [M+Na]+ 357.0885, found 357.0888.
Cell Culture
HuH6 cells,29 HuH7 (XenoTech JCRB0403) cells and HEK-293 cells (ATCC CRL-1573) were grown in Dulbecco’s Modified Eagle Medium (VWR #45000-304) supplemented with 10% FBS and 1% penicillin/streptomycin. HepG2 (ATCC HB-8065) were grown in RPMI-1640 Medium (VWR #10-040 CV) supplemented with 10% FBS, 1% sodium pyruvate and 1% penicillin/streptomycin. Hep3B cells (ATCC HB-8064) were grown in Eagle's Minimum Essential Medium (EMEM) (ATCC #30-2003) supplemented with 10% FBS and 1% penicillin/streptomycin.
Anti-Proliferative Assay
Compounds 4a-b, 7a-c, 10a-h, 13a-f, 17a-l, 21a-c, 23a-b, 24a-c, 26a-b, and 27a-c were initially dissolved in 100% DMSO. HuH6, HuH7, and HEK-293 cells were all seeded in 96-well plates at 2.5 x 104 cells/well. HepG2 and Hep3B cells were all seeded in 96-well plates at 3.5 x 104 cells/well. Cells were treated with increasing concentrations of all compounds for 72 h at 37 °C in a CO2 incubator. The MTT assay was performed with thiazolyl blue tetrazolium bromide (MTT, VWR #97062-380) dye as follows: 20 μL of MTT dye at a concentration of 5.4 mg /mL was added to each well, and then plates were incubated for 3 h at 37 °C in a CO2 incubator. Media was then removed and 100 μL of DMSO was added. Plates were then shaken at room temperature for 10 min to dissolve the purple-colored formazan crystals. After 10 min, the absorbance was measured using a Tecan SPARK10M spectrophotometer (Tecan Group Ltd., Mannedorf, Switzerland) at 570 nm. Each experiment had 6 repeats per treatment. Analysis and IC50 determination were performed using GraphPad Prism v6.0.
CETSA Analysis
HuH6 cells were seeded in a T175 flask and treated with an IC50 concentration of compound 13d in DMSO (0.25% DMSO final concentration), or 0.25% DMSO only for 5 h. Cells were harvested and divided into 6 aliquots, these individual aliquots were then subjected to heat treatment at either 50, 53, 56, 59, 62 or 65 °C. After heat treatment, cells were lysed with Radioimmunoprecipitation assay (RIPA) buffer and a protease/phosphatase inhibitor cocktail (ThermoFisher) on ice for 60 min at 4 °C. Protein concentrations were determined with the DC Protein Assay Kit (BioRad) per the manufacturer’s instructions. Equal amounts of protein (15 μg) were electrophoresed under reducing conditions, transferred to a PVDF membrane, and immunoblotted with the corresponding primary antibodies: anti-β-actin (Cell Signaling) and anti-Gankyrin (Cell Signaling). Membranes were incubated with an appropriate horseradish peroxidase-labeled secondary antibody (anti-Rabbit, Invitrogen), developed with WesternBright ECL chemiluminescent kit (Gel Company, San Francisco, CA), and visualized with an Omega LumG imaging system (Gel Company, San Francisco, CA).
Western blot analysis
HuH6 cells were seeded in 10 cm dishes and treated with varying concentrations of compounds 13d in DMSO (0.25% DMSO final concentration), or 0.25% DMSO only for 48 h. Cells were harvested in cold PBS and lysed with Radioimmunoprecipitation assay (RIPA) buffer and a protease/phosphatase inhibitor cocktail (ThermoFisher) on ice for 60 min at 4 °C. Protein concentrations were determined with the DC Protein Assay Kit (BioRad) per the manufacturer’s instructions. Equal amounts of protein (15 μg) were electrophoresed under reducing conditions, transferred to a PVDF membrane, and immunoblotted with the corresponding primary antibodies: anti-p53 (Cell Signaling), anti-Rb (Cell Signaling), anti-Cyclophilin (Cell Signaling), anti-Gankyrin (Cell Signaling). Membranes were incubated with an appropriate horseradish peroxidase-labeled secondary antibody (anti-Mouse, ThermoFisher Scientific; anti-Rabbit, Invitrogen), developed with WesternBright ECL chemiluminescent kit (Gel Company, San Francisco, CA), and visualized with an Omega LumG imaging system (Gel Company, San Francisco, CA).
Cell cycle analysis
HuH6 cells were seeded in 10 cm dishes and treated with 1.3 μM of compound 13d, in DMSO (0.25% DMSO final concentration), or 0.25% DMSO only for 72 h. Cells were harvested in cold PBS and normalized. The cells were then fixed in 70% ethanol and incubated at −20 °C overnight. Cells were then centrifuged, and the pellet was resuspended in 50 μL of PI/RNase kit (BD). The cells were then incubated for 30 min at 37 °C before performing flow and imaging cytometry analysis. Results were analyzed using FlowJo™ v10.8. One-way ANOVA was used to determine the statistical significance. A P-value < 0.05 was considered statistically significant.
Protein Expression and Purification of recombinant gankyrin and ANKRA2
The pQTEV-PSMD10 bacterial expression vector for gankyrin was obtained from Addgene (plasmid #31332). The plasmid was transformed into LOBSTR BL21 (DE3) Escherichia coli cell line on LB agar plates supplemented with 100 μg/mL ampicillin. Colonies were aseptically transferred into a sterile 250 mL baffled flask with 50 mL of autoclaved LB broth supplemented with 100 μg/mL ampicillin and placed in an incubator at 37 °C and 200 rpm overnight. Large scale production cultures with 1 L LB Broth and the appropriate antibiotics were inoculated with 25 mL of starter culture and grown at 37 °C until the OD600 reached 0.6–0.8. The cultures were then induced with 0.5 mM isopropyl thiogalactoside (IPTG) and grown at 37 °C and 200 rpm for 16 h. Cells were harvested by centrifugation with a Sorvall LYNX 4000 Superspeed Centrifuge with a Fiberlite F10-4x1000 LEX rotor (Thermo Fisher) at 6,000 rpm for 15 min at 4 °C. The cell pellets were then resuspended in wash buffer (25 mM Tris, 200 mM NaCl, 10 mM Imidazole, pH 8) supplemented with ProBlock™ Protease Inhibitor Cocktail (GoldBio) at 100 μL/mL. The cells were lysed using an EmulsiFlex-C5 homogenizer (Avestin), and the cell extract was clarified using a Beckman Coulter Optima L-100XP Preparative Ultracentrifuge with a Ti 70.1 rotor (Beckman Coulter) at 41,000 rpm for 40 min at 4 °C. The clarified supernatant was collected and passed through a 5.0 μm syringe filter (Thermo Fisher) before being loaded onto a 5 mL HisTrap FF column (Cytiva) on an ÄKTA Start FPLC (Cytiva). After 20 column volumes of wash buffer, gankyrin was eluted with elution buffer (25 mM Tris, 200 mM NaCl, 200 mM Imidazole, pH 8). The N-terminal poly-histidine tag was removed with TEV protease (40 μg/mL) and incubated for 1 h at room temperature on a platform shaker. The sample was then dialyzed to remove excess imidazole with 3.5 MWCO Spectra/Por 3 Dialysis Tubing (Thermo Fisher) in wash buffer at 4 °C gently mixing overnight. The sample was then passed through 5 mL HisTrap FF again to remove the His6-tag and TEV protease from the sample. If any protein contaminants remained, gel filtration chromatography using a Superdex 75 HiLoad column (Cytiva) was performed on an AKTA pure 25L FPLC system (Cytiva) equilibrated with wash buffer. Fractions containing pure gankyrin protein as assessed by SDS-PAGE were pooled, quantified by absorbance, aliquoted, flash frozen on liquid nitrogen, and stored at −80 °C.
The ANKRA2 bacterial expression vector was obtained from Addgene (plasmid #36904). The plasmid was transformed into transformed in a LOBSTR BL21 (DE3) Escherichia coli cell line on LB agar plates supplemented with 30 μg/mL kanamycin. The remainder of the ANKRA2 overexpression and purification was performed the same as described for gankyrin except for using kanamycin for selection during culture growth and protein overexpression.
Circular Dichroism
Circular dichroism (CD) experiments were collected on a Jasco J-815 Circular Dichroism Spectropolarimeter (JASCO Inc.). Each sample contained 10 μM of purified gankyrin or ANKRA2 in CD buffer (25 mM Tris, 50 mM NaCl, pH 7.4) alone or in the presence of 1 μM, 10 μM, and 100 μM compounds 10d or 13d. Samples were measured in a 1 mm quartz cuvette. CD spectra were collected from 195–260 nm, with six accumulation trials at 25 °C. All CD samples were performed in triplicate to ensure reproducibility.
Supplementary Material
Figure 6. Circular dichroism spectral analysis of gankyrin and ANKRA2 in the presence of compounds 10d (blue) and 13d (orange).
Protein alone samples (black) were saturated with increasing concentrations of compounds 10d and 13d (represented by a dark to light colorimetric scheme) to measure changes in α-helical content. Each protein was measured at 10 μM and introduced to compound concentrations ranging from 1 μM to 100 μM. Gankyrin samples are represented by circular markers while ANKRA2 samples are triangular markers. The CD spectra for each protein alone showed characteristic α-helical spectral minima at 208 and 222 nm. (A) In the presence of compound 10d, the CD spectra of gankyrin (left) depicted a reduction in secondary structure content as the drug concentration increased. Conversely, for ANKRA2 (right), the CD spectra showed no changes in secondary structure content. (B) For compound 13d, the CD spectra of gankyrin (left) indicated a decline in secondary structure content with increasing drug concentration. In contrast, the CD spectra of ANKRA2 (right) revealed no alterations in secondary structure content. Data shown is representative of triplicate runs.
Highlights.
Potent antiproliferative activity against certain liver cancers was exhibited
Minimal impact on potency from only replacing the propyl linker of cjoc42
Gankyrin stabilization was observed as a mechanism of inhibition
Gankyrin specific binding was demonstrated
Disruption of the proteasomal degradation pathway was demonstrated
Acknowledgement
This work was supported in part by NIH grant SC2GM139672 (A.M.) and R15GM126432 (D.E.S.). The authors also thank the College of Pharmacy and Health Sciences, the Department of Pharmaceutical Sciences, and the Office of Grants and Sponsored Research at St. John’s University, as well as the Gustaf H. Carlson School of Chemistry and Biochemistry at Clark University for their financial support of this research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data Availability
Data will be made available upon reasonable request.
References
- (1).Zhao X; Fu J; Xu A; Yu L; Zhu J; Dai R; Su B; Luo T; Li N; Qin W; Wang B; Jiang J; Li S; Chen Y; Wang H Gankyrin Drives Malignant Transformation of Chronic Liver Damage-Mediated Fibrosis via the Racl/JNK Pathway. Cell Death Dis. 2015, 6, e1751–e1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Jing H; Zhang G; Meng L; Meng Q; Mo H; Tai Y Gradually Elevated Expression of Gankyrin during Human Hepatocarcinogenesis and Its Clinicopathological Significance. Sci. Rep 2014, 4, 5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Higashitsuji H; Itoh K; Nagao T; Dawson S; Nonoguchi K; Kido T; Mayer RJ; Arii S; Fujita J Reduced Stability of Retinoblastoma Protein by Gankyrin, an Oncogenic Ankyrin-Repeat Protein Overexpressed in Hepatomas. Nat. Med 2000, 6, 96–99. [DOI] [PubMed] [Google Scholar]
- (4).Li J; Guo Y Gankyrin Oncoprotein: Structure, Function, and Involvement in Cancer. Curr. Chem. Biol 2010, 4, 13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Krzywda S; Brzozowski AM; Higashitsuji H; Fujita J; Welchman R; Dawson S; Mayer RJ; Wilkinson AJ The Crystal Structure of Gankyrin, an Oncoprotein Found in Complexes with Cyclin-Dependent Kinase 4, a 19 S Proteasomal ATPase Regulator, and the Tumor Suppressors Rb and P53. J. Biol. Chem 2004, 279, 1541–1545. [DOI] [PubMed] [Google Scholar]
- (6).D’Souza AM; Jiang Y; Cast A; Valanejad L; Wright M; Lewis K; Kumbaji M; Shah S; Smithrud D; Karns R; Shin S; Timchenko N Gankyrin Promotes Tumor-Suppressor Protein Degradation to Drive Hepatocyte Proliferation. Cell. Mol. Gastroenterol. Hepatol 2018, 6, 239–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Higashitsuji H; Liu Y; Mayer RJ; Fujita J The Oncoprotein Gankyrin Negatively Regulates Both P53 and RB by Enhancing Proteasomal Degradation. Cell Cycle 2005, 4, 1335–1337. [DOI] [PubMed] [Google Scholar]
- (8).Li H; Zhang J; Zhen C; Yang B; Feng L Gankyrin as a Potential Target for Tumor Therapy: Evidence and Perspectives. Am. J. Transl. Res 2018, 10, 1949–1960. [PMC free article] [PubMed] [Google Scholar]
- (9).Chapman AM; Rogers BE; McNaughton BR Characterization of the Binding Interaction between the Oncoprotein Gankyrin and a Grafted S6 ATPase. Biochemistry 2014, 53, 6857–6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Li J; Knobloch TJ; Kresty LA; Zhang Z; Lang JC; Schuller DE; Weghorst CM Gankyrin, A Biomarker for Epithelial Carcinogenesis, Is Overexpressed in Human Oral Cancer. Anticancer Res. 2011, 31, 2683–2692. [PubMed] [Google Scholar]
- (11).Kashyap D; Varshney N; Parmar HS; Jha HC Gankyrin: At the Crossroads of Cancer Diagnosis, Disease Prognosis, and Development of Efficient Cancer Therapeutics. Adv. Cancer Biol. - Metastasis 2022, 4, 100023. [Google Scholar]
- (12).Kanabar D. Small Molecule Gankyrin Inhibition as a Therapeutic Strategy for Breast and Lung Cancer. PhD Thesis, St. John's University, Queens, NY, USA, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Hwang J-A; Yang H-M; Hong D-P; Joo S-Y; Choi Y-L; Park J-H; Lazar AJ; Pollock RE; Lev D; Kim SJ Gankyrin Is a Predictive and Oncogenic Factor in Well-Differentiated and Dedifferentiated Liposarcoma. Oncotarget 2014, 5, 9065–9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Mayer RJ; Fujita J Gankyrin, the 26 S Proteasome, the Cell Cycle and Cancer. Biochem. Soc. Trans 2006, 34, 746–748. [DOI] [PubMed] [Google Scholar]
- (15).Zamani P; Matbou Riahi M; Momtazi-Borojeni AA; Jamialahmadi K Gankyrin: A Novel Promising Therapeutic Target for Hepatocellular Carcinoma. Artif. Cells Nanomedicine Biotechnol 2018, 46, 1301–1313. [DOI] [PubMed] [Google Scholar]
- (16).Dawson S; Apcher S; Mee M; Mayer RJ; Higashitsuji H; Baker R; Uhle S; Dubiel W; Fujita J Gankyrin Is an Ankyrin-Repeat Oncoprotein That Interacts with CDK4 Kinase and the S6 ATPase of the 26 S Proteasome. J. Biol. Chem 2002, 277, 10893–10902. [DOI] [PubMed] [Google Scholar]
- (17).Higashitsuji H; Higashitsuji H; Itoh K; Sakurai T; Nagao T; Sumitomo H; Masuda T; Dawson S; Shimada Y; Mayer RJ; Fujita J The Oncoprotein Gankyrin Binds to MDM2/HDM2, Enhancing Ubiquitylation and Degradation of P53. Cancer Cell 2005, 8, 75–87. [DOI] [PubMed] [Google Scholar]
- (18).Nanaware PP; Ramteke MP; Somavarapu AK; Venkatraman P Discovery of Multiple Interacting Partners of Gankyrin, a Proteasomal Chaperone and an Oncoprotein-Evidence for a Common Hot Spot Site at the Interface and Its Functional Relevance: Protein Interaction Network of a Hub Oncoprotein. Proteins Struct. Funct. Bioinforma 2014, 82, 1283–1300. [DOI] [PubMed] [Google Scholar]
- (19).Wang C; Cheng L Gankyrin as a Potential Therapeutic Target for Cancer. Invest. New Drugs 2017, 35, 655–661. [DOI] [PubMed] [Google Scholar]
- (20).Dawson S; Higashitsuji H; Wilkinson AJ; Fujita J; Mayer RJ Gankyrin: A New Oncoprotein and Regulator of pRb and P53. Trends Cell Biol. 2006, 16, 229–233. [DOI] [PubMed] [Google Scholar]
- (21).Man J-H; Liang B; Gu Y-X; Zhou T; Li A-L; Li T; Jin B-F; Bai B; Zhang H-Y; Zhang W-N; Li W-H; Gong W-L; Li H-Y; Zhang X-M Gankyrin Plays an Essential Role in Ras-Induced Tumorigenesis through Regulation of the RhoA/ROCK Pathway in Mammalian Cells. J. Clin. Invest 2010, 120, 2829–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).D’Souza AM; Cast A; Kumbaji M; Rivas M; Gulati R; Johnston M; Smithrud D; Geller J; Timchenko N Small Molecule Cjoc42 Improves Chemo-Sensitivity and Increases Levels of Tumor Suppressor Proteins in Hepatoblastoma Cells and in Mice by Inhibiting Oncogene Gankyrin. Front. Pharmacol 2021, 12, 580722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Li H; Fu X; Chen Y; Hong Y; Tan Y; Cao H; Wu M; Wang H Use of Adenovirus-Delivered siRNA to Target Oncoprotein p28GANK in Hepatocellular Carcinoma. Gastroenterology 2005, 128, 2029–2041. [DOI] [PubMed] [Google Scholar]
- (24).Qian Y; Chen Y; Yang W; Fu J; Cao J; Ren Y; Zhu J; Su B; Luo T; Zhao X; Dai R; Li J; Sun W; Wu M; Feng G; Wang H p28GANK Prevents Degradation of Oct4 and Promotes Expansion of Tumor-Initiating Cells in Hepatocarcinogenesis. Gastroenterology 2012, 142, 1547–1558. [DOI] [PubMed] [Google Scholar]
- (25).Chattopadhyay A; O’Connor CJ; Zhang F; Galvagnion C; Galloway WRJD; Tan YS; Stokes JE; Rahman T; Verma C; Spring DR; Itzhaki LS Discovery of a Small-Molecule Binder of the Oncoprotein Gankyrin That Modulates Gankyrin Activity in the Cell. Sci. Rep 2016, 6. 23732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Nakamura Y; Nakano K; Umehara T; Kimura M; Hayashizaki Y; Tanaka A; Horikoshi M; Padmanabhan B; Yokoyama S Structure of the Oncoprotein Gankyrin in Complex with S6 ATPase of the 26S Proteasome. Structure 2007, 15, 179–189. [DOI] [PubMed] [Google Scholar]
- (27).Kanabar D; Farrales P; Gnanamony M; Almasri J; Abo-Ali EM; Otmankel Y; Shah H; Nguyen D; El Menyewi M; Dukhande VV; D’Souza A; Muth A Structural Modification of the Aryl Sulfonate Ester of Cjoc42 for Enhanced Gankyrin Binding and Anti-Cancer Activity. Bioorg. Med. Chem. Lett 2020, 30, 126889. [DOI] [PubMed] [Google Scholar]
- (28).Kanabar D; Farrales P; Kabir A; Juang D; Gnanmony M; Almasri J; Torrents N; Shukla S; Gupta V; Dukhande VV; D’Souza A; Muth A Optimizing the Aryl-Triazole of Cjoc42 for Enhanced Gankyrin Binding and Anti-Cancer Activity. Bioorg. Med. Chem. Lett 2020, 30, 127372. [DOI] [PubMed] [Google Scholar]
- (29).Kanabar D; Goyal M; Kane EI; Chavan T; Kabir A; Wang X; Shukla S; Almasri J; Goswami S; Osman G; Kokolis M; Spratt DE; Gupta V; Muth A Small-Molecule Gankyrin Inhibition as a Therapeutic Strategy for Breast and Lung Cancer. J. Med. Chem 2022, 65, 8975–8997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Ozaki T; Nakagawara A Role of P53 in Cell Death and Human Cancers. Cancers 2011, 3, 994–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Engeland K. Cell Cycle Regulation: P53-P21-RB Signaling. Cell Death Differ. 2022, 29, 946–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Giacinti C; Giordano A RB and Cell Cycle Progression. Oncogene 2006, 25, 5220–5227. [DOI] [PubMed] [Google Scholar]
- (33).He M; Zhao M; Shen B; Prise KM; Shao C Radiation-Induced Intercellular Signaling Mediated by Cytochrome-c via a P53-Dependent Pathway in Hepatoma Cells. Oncogene 2011, 30, 1947–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Rodríguez-Hernández MA; Chapresto-Garzón R; Cadenas M; Navarro-Villarán E; Negrete M; Gómez-Bravo MA; Victor VM; Padillo FJ; Muntané J Differential Effectiveness of Tyrosine Kinase Inhibitors in 2D/3D Culture According to Cell Differentiation, P53 Status and Mitochondrial Respiration in Liver Cancer Cells. Cell Death Dis. 2020, 11, 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Waldherr M; Mišík M; Ferk F; Tome J; Žegura B; Filipič M; Mikulits W; Mai S; Haas O; Huber WW; Haslinger E; Knasmüller S Use of HuH6 and Other Human-Derived Hepatoma Lines for the Detection of Genotoxins: A New Hope for Laboratory Animals? Arch. Toxicol 2018, 92, 921–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Golub T; Dou G; Zeng C; Becker JY One-Pot Anodic Conversion of Symmetrical Bisamides of Ethylene Diamine to Unsymmetrical Gem -Bisamides of Methylene Diamine. Org. Lett 2019, 21, 7961–7964. [DOI] [PubMed] [Google Scholar]
- (37).Habash M; Taha MO Ligand-Based Modelling Followed by Synthetic Exploration Unveil Novel Glycogen Phosphorylase Inhibitory Leads. Bioorg. Med. Chem 2011, 19, 4746–4771. [DOI] [PubMed] [Google Scholar]
- (38).Kubicova L; Waisser K; Kunes J; Kralova K; Odlerova Z; Janota J; Svoboda Z Synthesis of N,N’-Diarylalkanediamides and Their Antimycobacterial and Antialgal Activity. Molecules 2000, 5, 714–726. [Google Scholar]
- (39).Zamanloo MR; Karimi MH; Imanzadeh GH Highly Compact Co-Poly(Amide-Imide)s from Polycondensation of an Imide-Modified Derivative of 1-Aspartic Acid. E-Polym. 2014, 14, 417–425. [Google Scholar]
- (40).Aziz H; Saeed A; Jabeen F; Ullah N; Rehman AU Synthesis, Characterization, in Vitro Biological and Molecular Docking Evaluation of N,N’-(Ethane-1,2-Diyl)Bis(Benzamides). J. Iran. Chem. Soc 2021, 18, 2425–2436. [Google Scholar]
- (41).Guarda L; Parra N; Chávez MS; Belmar J; Jiménez CA; Pasán J; Ruiz-Pérez C Synthesis and Characterization of Bis- and tris-(4-carboxybenzoyl)-alkaneamines. J Chil Chem Soc 2012, 3, 1305–1308. [Google Scholar]
- (42).Flick AC; Leverett CA; Ding HX; McInturff E; Fink SJ; Helal CJ; DeForest JC; Morse PD; Mahapatra S; O’Donnell CJ Synthetic Approaches to New Drugs Approved during 2018. J. Med. Chem 2020, 63, 10652–10704. [DOI] [PubMed] [Google Scholar]
- (43).Song X; Liu X; Yu W; Jin Y Amide-Assisted Rearrangement of Hydroxyarylformimidoyl Chloride to Diarylurea. Molecules 2021, 26, 6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Zhao Y; Guo X; Si Z; Hu Y; Sun Y; Liu Y; Ji Z; You J Hydrosilane-Assisted Synthesis of Urea Derivatives from CO 2 and Amines. J. Org. Chem 2020, 85, 13347–13353. [DOI] [PubMed] [Google Scholar]
- (45).Yang H-T; Tan Y-C; Yang Y; Sun X-Q; Miao C-B Cu(OAc) 2 -Mediated Reaction of C60 with Ureas for the Preparation of Fulleroimidazolidinones. J. Org. Chem 2016, 81, 1157–1163. [DOI] [PubMed] [Google Scholar]
- (46).Howbert JJ; Grossman CS; Crowell TA; Rieder BJ; Harper RW; Kramer KE; Tao EV; Aikins J; Poore GA Novel Agents Effective against Solid Tumors: The Diarylsulfonylureas. Synthesis, Activities, and Analysis of Quantitative Structure-Activity Relationships. J. Med. Chem 1990, 33, 2393–2407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available upon reasonable request.