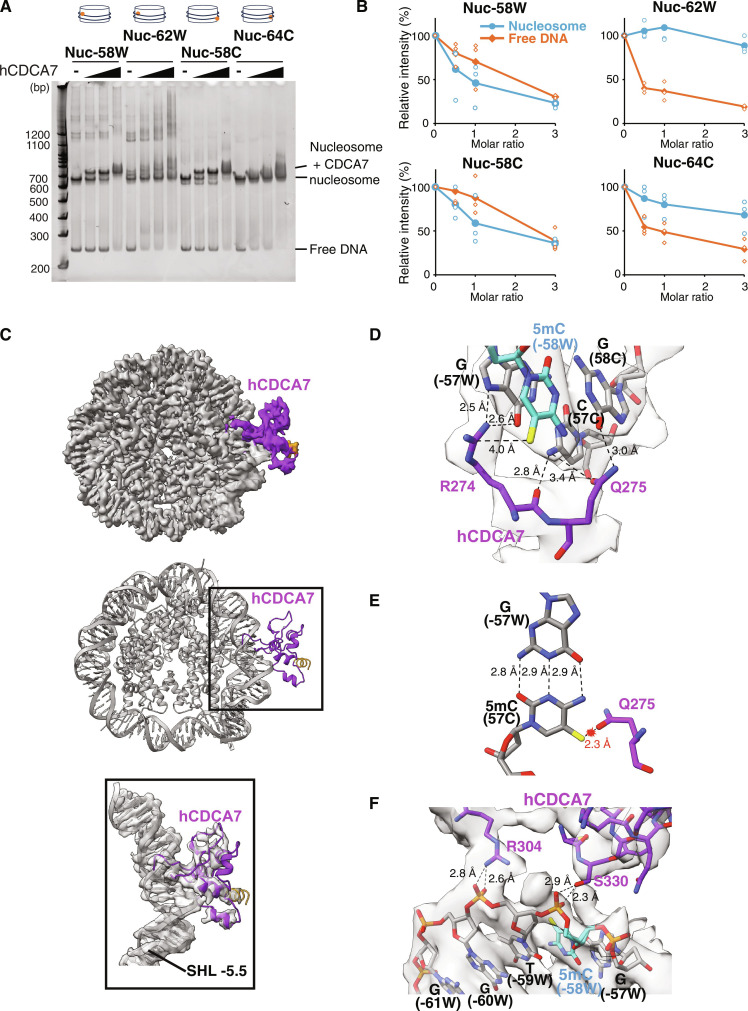
Fig. 4. Cryo-EM structure of hCDCA7 bound at NCP.
(A) EMSA analyzing the interaction of hCDCA7264–371 C339S with nucleosomes carrying hemimethylated CpG at the indicated positions. (B) Quantification of the free DNA (orange) and nucleosome signal (blue) detected by EMSA upon the addition of hCDCA7264–371 C339S relative to the signal detected in the absence of CDCA7. Line graph shows the average from n = 3 independent experiments. (C) A composite cryo-EM map (top) and the structure of hCDCA7264–371 C339S (middle) bound to the nucleosome harboring a 5mC at the Watson strand, position -58 (Nuc -58W). The map corresponding to CDCA7 is colored purple, conserved C-terminal helix indicated in orange. Overlay of atomic model of hCDCA7264–371 C339S on the cryo-EM map (bottom). (D) A structure of hCDCA7264–371 C339S bound to the nucleosome (Nuc -58W). Key residues for the selective recognition of hemimethylation, R274 and Q275, are shown as purple stick model. The methyl group of 5mC is shown in yellow. (E) Predicted steric clash between Q275 and the methyl group at the Crick strand, 5mC position 57 in a nucleosome harboring a fully methylated CpG dyad (5mC -58 W/5mC 57C) (F) A structure of hCDCA7264–371 C339S bound to the nucleosome (Nuc -58 W). Residues contacting the phosphate backbone of DNA are shown.