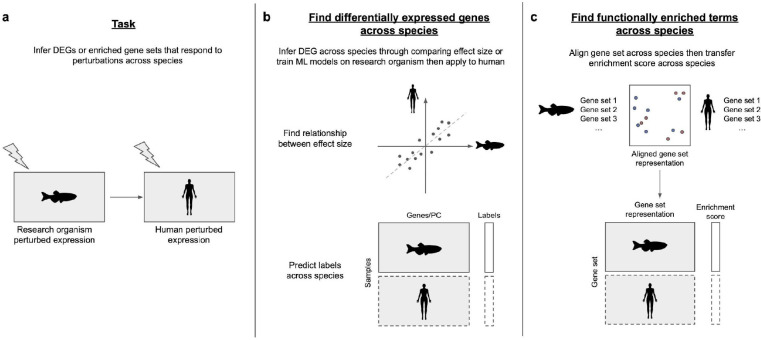
Figure 4: How to infer perturbed transcriptomes across species.
(a) The objective is to infer potential perturbed genes or gene sets in humans based on experimental perturbation results in research organisms. (b) To infer DEGs across species, FIT [89] uses linear models to describe relationships between the effect sizes of DEGs in humans and research organisms with matching conditions. Other approaches develop ML models based on research organisms to predict perturbed expression in humans. (c) To infer enriched gene sets across species, XGSEA [90] aligns gene set representations across species, train models in research organisms, and then infer enrichment scores of gene sets across species.